* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Pathway analysis
Survey
Document related concepts
Transcript
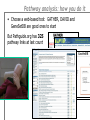
Pathway analysis Daniel Hurley Pathway analysis: summary A popular buzzword… but what does it mean? How do you do it? How do you interpret the results? First What we mean by ‘pathway analysis’ • A ‘pathway’ implies causation, but don’t be fooled! What we mean by ‘pathway analysis’ • A ‘pathway’ implies causation, but don’t be fooled! • Most ‘pathway analysis’ actually identifies groups of functionally similar transcripts. • Louis’ example: (http://gather.genome.duke.edu/) What we mean by ‘pathway analysis’ • A useful paper…. But the conclusion is: lots of tools, some quite different approaches! What we mean by ‘pathway analysis’ • Pathway analysis tools like GATHER, DAVID, and GeneSetDB typically rely on enrichment analyses to tell us things. • This set of techniques asks the question ‘of this set of genes, how many share any particular function, and is that more than we would expect by chance?’ • Example: the top 200 most differentially-expressed genes by some ranking (e.g. adjusted p-value) • Determination of ‘by chance’ is usually done using a permutation (= Monte Carlo) approach • Other ‘pathway analyses’ involve signatures of groups of transcripts (e.g. using Principal Component Analysis) What we mean by ‘pathway analysis’ But what do we mean by a ‘function’? Lots of things: Protein function Hypothetical protein function Chromosomal location Metabolic pathway association Disease association Daunting The key point What we mean by ‘pathway analysis’ • Pathway analysis can identify common features present in a group of transcripts • What the output means depends on the specific biology under study • No such thing really as a ‘general’ pathway analysis • A good place to start is by finding papers relevant to the specific biology What can you do with it? Some answers: Get a general picture of the active functions in a condition (vs. control) Identify conditions with similar functions Investigate the functions associated with a particular gene Investigate whether a particular function is active in a condition Differentiate conditions by their active functions Next Pathway analysis: how you do it • Begin with a list of transcripts of interest Pathway analysis: how you do it • Choose a web-based tool: GATHER, DAVID and GeneSetDB are good ones to start But Pathguide.org has 325 pathway links at last count Pathway analysis: how you do it • Enter the list of transcripts: with most tools, you will either paste in gene names or identifiers, or upload a file Finally Pathway analysis: interpreting results • Basic tools will produce ranked lists of the most ‘enriched’ categories: GATHER Pathway analysis: interpreting results • More sophisticated ones will produce ‘network’ diagrams DAVID Ingenuity Pathways But the interpretation of these is Analysis rather subjective Summary • Pathway analysis should probably be called information enrichment analysis – a more accurate term • Used prudently, it is a useful tool for exploring the functional landscape of an experiment • To make it meaningful, you need to interpret the results in the context of the specific biology under study • There are a lot of web-based tools; start with one which is current and produces a result you value • To start, you need a set of (transcripts) of interest • To present the results, you can use a simple table, or a more complex ‘network’ diagram • Risk: false-positives are very difficult to identify, and with enough data you can link any molecular species to any other species FIN Any questions?