* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download The Effect of the BAF Chromatin Regulator in Heart Valve Maturation
Electrocardiography wikipedia , lookup
Rheumatic fever wikipedia , lookup
Aortic stenosis wikipedia , lookup
Hypertrophic cardiomyopathy wikipedia , lookup
Pericardial heart valves wikipedia , lookup
Arrhythmogenic right ventricular dysplasia wikipedia , lookup
Lutembacher's syndrome wikipedia , lookup
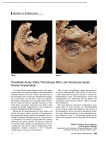
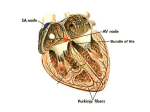
THE EFFECT OF THE BAF CHROMATIN REGULATOR IN HEART VALVE MATURATION OF MUS MUSCULUS Kelsey Ward Kryn Stankunas, P.I. Brynn Simek, Mentor CONGENITAL HEART DISEASE (CHD) • Most common type of birth defect •1 in 125 births EPIGENETICS AND CHROMATIN REMODELING VALVULOGENESIS Chromatin Remodeling by the BAF Complex Brg1 VALVULOGENESIS BRG1 DELETION IN ALL ENDOCARDIAL CELLS IS EMBRYONIC LETHAL Stankunas, K. Methodology ♂ SCL-Cre-ER(T); Brg1F/F ♀ Brg1 F/F Genotype embryos Imaging whole embryos Characterize valve defects and consider new time points and crosses • Check Plugs • Administer Tamoxifen (2x40mg/kg) • Harvest embryos • • • • • Preserve in PFA Transfer to PBS Dehydrate to 100% EtOH Replace with xylenes Replace with paraffin and fix in blocks • Section at 7 microns • H&E stain • Image sections VALVULOGENESIS TAMOXIFEN ADMINISTRATION AT E12.5 A B C D IS BRG1 ACTUALLY DELETED? Pulmonic Valve Mitral Valve HOW QUICKLY DOES DELETION OCCUR? VENTRICULAR SEPTAL DEFECTS 79.8 3.5 HOW EFFICIENT IS DELETION? CONCLUSIONS AND FUTURE DIRECTIONS Brg1 deletion has no phenotypic effect on later valve maturation. Characterization of Scl-cre line and optimization of Tamoxifen dosage. Earlier Brg1 deletion leads to cushion defects and ventricular defects. Allowance of a longer growth period before harvesting. Describing the molecular and cellular basis for the phenotypes. ACKNOWLEDGEMENTS Brynn Simek, Mentor Kryn Stankunas, P.I. Stankunas Lab •Alex •Andrew •Fern •Matt •Scott SPUR