* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download NGS: Coming to a lab near you!
Survey
Document related concepts
Transcript
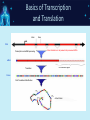
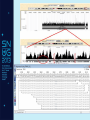
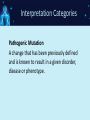
NGS: Coming to a Lab Near You! An Introduction to Next Generation Sequencing (NGS) SNUG 2013 Laurel Estabrooks, PhD, FACMG VP Genetics Business Development SCC Soft Computer What is “DNA Sequencing”? • DNA sequencing involves the use of various methods for determining the order of the nucleotide bases — adenine, cytosine, guanine, and thymine — in a molecule of DNA. Exon Intron Gene Exon DNA Basics • Bases - In molecular biology and genetics, two nucleotides on opposite complementary DNA or RNA strands that are connected via hydrogen bonds are called a base pair. Adenine (A) forms a base pair with thymine (T) and guanine (G) forms a base pair with cytosine (C). In RNA, thymine is replaced by uracil (U), and therefore bonds to Adenine (A). • Genetic code - the set of rules by which information encoded in genetic material (DNA or mRNA sequences) is translated into proteins (amino acid sequences) by living cells. It is a triplet code in that three nucleotides (a codon) determine particular amino acids. Basics of Transcription and Translation DNA mRNA Protein Basics of Transcription and Translation Intron Exon DNA Transcription and mRNA processing Intron information is not passed onto processed mRNA mRNA Translation Un-translated region Protein Post-Translational Modification Active Protein What is Next Generation Sequencing? • 1st Generation = Sanger Sequencing – 2 reads (forward & reverse) • 2nd Generation = Next Generation Sequencing – Millions of reads • 3rd Generation = Single Molecule Sequencing What is Next Generation Sequencing? What is Next Generation Sequencing? Major computations performed with NGS data • Data assembly with base calling at the level of individual reads • Alignment of the assembled sequence to a reference sequence • Variant calling NGS Alignment Multiple, fragmented sequence reads must be assembled together on the basis of their overlapping areas. NGS Technology Terminology • Read length - the average number of contiguous nucleotide bases in a polynucleotide sequence that are produced by a particular sequencing instrument (14-400) • Coverage – Number of times a nucleotide base is read (# followed by X: 300X) • Call – determination of a given base or base sequence by a sequencing instrument • Call Quality – accuracy of the call determination Base Calling Accuracy Q Scores • Base calling accuracy often measured by the Phred Quality Score (Q score) which assesses the accuracy of a sequencing platform. It indicates the probability that a given base is called incorrectly by the sequencer. • Logorithmic calculation • Q10 1/10 error rate • Q20 1/100 error rate • Q30 1/1000 error rate Example: Phred score of 30 (Q30) = probability of an incorrect base call 1 in 1000 times • Low Q scores can result in an increase in false positive variant calls There are multiple types of DNA changes including: Substitution Duplication Inversion Translocation Insertion/Deletion (Indel) SNPs - Single Nucleotide Polymorphisms – Substitution change in more than 1% of the population – Considered a common variant CNVs - Copy Number Variations – Sections of DNA bases in our genomes that are commonly copied many times over – Number of copies may vary from person to person Applications in Microbiology • Identifying the species of an isolate • Defining its properties, such as resistance to antibiotics and virulence • Monitoring the emergence and spread of bacterial pathogens Phylogenic Map NGS & Microbiology Case Study The NHS Rosie Hospital in Cambridge manages around 6,000 baby deliveries each year. All infants in its special care baby unit are screened for MRSA when admitted, and for every week while in the unit. This routine screening picked up MRSA in 12 infants. The Lancet Infectious Diseases, Volume 13, Issue 2, Pages 130 - 136, February 2013 NGS & Microbiology Case Study The following was performed: • Bacteria was cultured from swabs and plated on selective media. • Antimicrobial susceptibility was tested against an array of antibiotics. • Sequencing libraries were prepared from each MRSA isolate, and amplified. • Whole genome sequencing was performed using the Illumina MiSeq sequencer. The Lancet Infectious Diseases, Volume 13, Issue 2, Pages 130 - 136, February 2013 NGS & Microbiology Case Study • All affected infants were treated • Unit was sanitized The Lancet Infectious Diseases, Volume 13, Issue 2, Pages 130 - 136, February 2013 NGS & Microbiology Case Study Results • 14/17 infants had a new sequence type ST2371 • Only 20 SNPs varied among the 14 ST2371 isolates • ST22 is common MRSA sequence type in UK • ST2371 differs from ST22 isolate by an average of 550 SNPs NGS & Microbiology Case Study • Short hiatus from outbreaks • Another outbreak • Tested all SCBU personnel The Lancet Infectious Diseases, Volume 13, Issue 2, Pages 130 - 136, February 2013 Case Study Analysis Case Study: NGS Benefit • Identification of asymptomatic carrier causing re-infections • Upon treatment of carrier, no further outbreaks The Lancet Infectious Diseases, Volume 13, Issue 2, Pages 130 - 136, February 2013 NGS in Human Genetics Next Generation Sequencing Targeted Panel Whole Exome Whole Genome -smaller -targets entire coding region -targets entire genome -incidental findings -incidental findings target region -no incidental findings Incidental Findings • Findings not associated with the original trigger for the testing • Currently under debate regarding whether to report • Recent guidelines published from American College of Medical Genetics and Genomics Next Generation Sequencing Test Ordering Test Interpretation • Unknown diagnosis • Suspected diagnosis of disease with mutational heterogeneity • Available variant data • Patient clinical presentation • Co-segregation of variant with clinical issue in family Interpretation Categories Pathogenic Mutation A change that has been previously defined and is known to result in a given disorder, disease or phenotype. Interpretation Categories Probably/Possibly Pathogenic Not a defined change, but there is additional evidence based on – the gene involved, – the gene position, – the type of the variation, or – the family history that lends greater likelihood that this could indeed be the origin of the patient’s clinical presentation/disorder. How do you determine a variant is possibly/probably pathogenic? Use algorithms to assess how variation within a known gene would theoretically impact gene integrity, gene translation, or protein formation Example online tools: – PolyPhen 2 http://genetics.bwh.harvard.edu/pph2/ (Polymorphic Phenotyping - predicts loss of function). PolyPhen-2 is a tool which predicts possible impact of an amino acid substitution on the structure and function of a human protein using straightforward physical and comparative considerations. – SIFT http://sift.jcvi.org/ (Sorting Intolerant From Tolerant, just computes >4.55 Mb deletions) SIFT predicts whether an amino acid substitution affects protein function. Interpretation Categories Variant of Unknown Significance • Do not know the significance at this time • Incidence WGS>WES Example of Result Tables Excerpt of Interpretation Illustrating Interpretation Categories Questions?