* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download Biostatistics Quantitative Data • Descriptive Statistics • Statistical
Foundations of statistics wikipedia , lookup
Bootstrapping (statistics) wikipedia , lookup
Taylor's law wikipedia , lookup
Categorical variable wikipedia , lookup
History of statistics wikipedia , lookup
Resampling (statistics) wikipedia , lookup
Student's t-test wikipedia , lookup
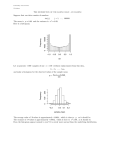
Biostatistics Quantitative Data Quantitative Data This course will focus on the analysis of quantitative data which is encountered in many areas of experimental research. Data may roughly be grouped into 3 groups : • Quantitative data : sperm concentration (mill/ml), height in cm, level of hormones (measured on a continuous scale). • Qualitative data : sex, race, work, groupings of quantitative data (high/medium/low). • Survival data : length of waiting time for some event. For some individuals, however, the event is never recorded. These individuals are censored and this makes some particular methods necessary. • Descriptive Statistics • Statistical Models • One-sample and Two-Sample Tests • Introduction to SAS-ANALYST • T- and Rank-Tests using ANALYST Thomas Scheike We will concentrate on quantitative data and describe : • Descriptive techniques. (Histograms, scatter-plots, means, standard deviation, quantiles, percentiles, ...) • Non-parametric methods. These are based on ranks of data, and may be used for one-sample tests, two-sample tests (paired and un-paired), one-sided analysis of variance and computation of measures of association (Spearman correlation). • Regression analysis techniques for normally distributed residuals. These techniques include : t-test (paired and un-paired such), analysis of variance (one- and two-sided), regression analysis, multiple regression analysis, analysis of covariance) We do, however, not discuss how to deal with repeated measures where subjects are followed and measured repeatedly. When repeated measures are encountered they may often be reduced to just one summary number for each subject and thereby analysed by techniques dealt with in this course. 1 2 Descriptive Statistics We consider data on sperm concentration (mill/ml) on two groups of people in a study. One group are members of an association that promotes the development of organic agriculture (n=55), and another group of workers are from a major Scandinavian airline carrier (n=141). How these data were collected is very important if we want to conclude more generally from the data. The data for both groups must be representative for the members of organic agriculture associations and airline workers. This must be very carefully validated, but for now we believe that this is the case. The Histogram The histogram is a different and better summary, it describes the distribution of the sperm concentrations for the two groups : Airline 40 Frequency 0 Drawing the data is the most important part of the statistical analysis : 0 5 20 10 Frequency 15 60 20 80 Organic farmers 0 100 200 300 400 0 100 200 300 400 sas 400 eco 200 100 height × width = Frequency in group if bars all have the same width this is not important. A difficulty is to decide the width of the bars. Here are two different histograms: 0 Histogram of conc Histogram conc 0.0 4 0.0 3 0 100 200 300 400 0 conc 3 of 2.0 Density 1.8 0.0 2 1.6 Group 0.0 1 1.4 0.0 0 1.2 Density 1.0 0.0 0 0.0 2 0.0 4 0.0 6 0.0 8 0.010 0.012 sperm concentration 300 A histogram shows how the data is distributed, i.e., we can find out how many men that have a sperm count lower that 100 mill/ml, say. For the Airline people this is 110 (141) men and the organic farmers have 35 (55) under 100 mill/ml. It is made by grouping of the sperm concentrations and then deciding the height of each bar such that: 100 200 conc 4 300 400 The Histogram The histogram describes the variability of the data. And we can approximate the chance that a data-point is below some limit, above some limit or between two limits by calculating the area of the histogram in the appropriate area : Percentiles Frequency 60 80 Histogram of conc 40 Area is = chance (Frequency / number ) 0 0.008 20 Histogram of conc 0 100 200 300 400 0.006 conc 0.000 0.002 0.004 Density To describe the histogram we may find the data value for which 50 % of the data is above or equal to and 50 % is below or equal to, this is the median. After ordering the data in size the median is the value in the middle of the data, for an even number of data points the median is the average of the two middle values : 0 100 200 300 1 4 6 8 9 ∼ median = 6 400 conc What is the probability of seeing a sperm concentration less than 40, say, from a randomly chosen man among our men in the study. 1 4 6 7 8 9 ∼ median = (6 + 7)/2 = 6.5 Similarly the 25% percentile (quantile) is the data point for which at least 25% of the data points have a lower or equal value and at least 75 % have a higher or equal value : 1 4 6 8 9 ∼ 25%percentile = 4 1 4 6 7 8 9 ∼ 25%percentile = 4 Find an approximate median in the histogram ? 5 6 Simple Summary Statistics We can calculate the mean (average) and standard-deviation for the two groups : n 1 X xi , x̄ = n i=1 n 1 X Variance = (xi − x̄)2, n − 1 i=1 and v u u n 1 X u SD = t (xi − x̄)2 n − 1 i=1 The mean describes the midpoint of the data, and the standard deviation the spread of the data. These number may always be calculated. Symmetric distributions are well characterized by these numbers, whereas a skewed distribution will not be well described. The Histogram The histogram based on the data is an approximation of the population the data is a representative sample from. A particularly nice histogram curve is the normal distribution : 0.2 0.1 normal density 0.3 0.4 Normal Distribution 0.0 0.4 Normal Distribution −2 0 2 4 7 of conc Histogram of conc^0.3 0.4 Histogram 0.3 0.2 Density 0.1 0.0 If a distribution does not appear symmetric one should instead compute median and various percentiles (25 % and 75 %, say) or give the range of the data (largest and smallest value). For the Sperm data the spermconcentration was 77 (77) (mean (SD)), the median and range was 56 and [0,402], respectively. What numbers are best suited to describe how the sperm concentration varies ?? 80 4 60 2 x Frequency 0 40 −2 20 0.0 −4 which is a good approximation to many symmetric histograms. Some properties of the normal curve is : • The normal curve is symmetric around its mean. • It is completely described by its mean and SD. By saying that data is normally distributed we mean that the histogram of the data is close (well approximated) to the normal curve. Sometimes a transformation of the data is necessary to make this true 0 0.2 x 0.1 normal density 0.3 −4 0 100 200 300 400 0 conc 1 2 3 4 conc^0.3 8 5 6 Histogram of height 0.006 0.000 0.2 0.002 0.4 0.004 pnorm(x) 0.6 Density 0.008 0.8 0.010 1.0 There are tables of the standard normal distribution which has mean=0 and SD=1, and the area between two values for any other normal curve can be found using this table by converting values to standard scores. Example : The height of Danish women are approximately normal with mean 165cm and standard deviation 30cm. If a woman is chosen at random what is the chance that she is lower than 180 cm. Standard score = (180-165)/30 = 0.5, i.e., 180 is 0.5 standard deviations above the mean. The chance of being less than 0.5 in a standard normal is 0.65 Is it a reasonable statistical model ?? What is the chance of a randomly chosen woman is between 190 and 175? Convert to standard scores = 0.83, 0.33 0.012 The Normal Distribution Similarly, to how we use the histogram, based on the normal curve we can work out how the data is distributed. The normal curve satisfies that : • 50 % of the area is under the mean. • 95 % of the area is between [mean - 1.96 SD, mean + 1.96 SD]. • 68 % of the area is between [mean - 1 SD, mean + 1 SD]. • 2.5 % of the area is between [−∞, mean − 1.96SD]. 150 0.0 100 200 250 height −4 −2 0 2 4 x The figure gives the cumulative distribution, i.e., what percent of the distribution is below a given value. The statement may formally be written as : P (X < 0.83) = 0.80; P (X < 0.33) = 0.63and P (0.33 < X < 0.83) = P (X < 0.83) − P (X < 0.33) = 0.80 − 0.63 This is based on the following precise statement about standard scores. With Z normal with mean µ and variance σ 2 it follows that (Z − µ)/σ is standard normal. 9 10 Distributions We often draw histogram curves to show how the data is distributed (is varying). How does these two histograms differ from the normal curve : Example: Suppose that the sperm-concentration in the Danish population is right skewed : Standard Log Normal Distribution Normal Distribution meanlog=3,sdlog=1 0.4 0.5 0.6 0.0 0 0.0 5 0.010 0.015 0.020 0.025 0.030 Log 100 200 300 400 0.2 0.3 0 0.0 0.1 If we draw 50 men at random from this distribution we get the following numbers : 2 4 6 8 10 Histogram for sample of 50 10 5 The first distribution is right skewed. i.e. data from this distribution contains some very high values. Frequency 15 20 0 0 Multi−Modal Histogram of Distribution c(x1, x2) 50 100 150 0 20 40 60 Frequency 80 100 120 140 0 2 4 6 8 10 12 calculations give mean=27, SD=29, median=17, range=2,250 Now, drawing again gives that : calculations give mean=34, SD=27, median=21, range=4,153 and again : calculations give mean=53 , SD=115 , median= 16, range=2,287 and again : calculations give mean=26 , SD=31 , median=20 , range=2,258 c(x1, x2) This other curve have several modes (multi-modal). 11 12 Example cont’d : Looking at concentrations on log-scale the population is distributed as follows : Distribution mean=3,sd=1 0.4 Normal Descriptive Statistics : Summary • The histogram shows how the data is distributed, i.e., how it is varying. 0.2 • The normal distribution is a histogram curve that is a good approximation to many histograms. 0.0 0.1 dnorm(3 + x, 3, 1) 0.3 • The area of the histogram represents frequency. 0 2 4 3 + 6 x Drawing 50 men randomly from the population gives the following histogram : calculations give mean=2.9 , SD=0.99 , median=2.8 , Histogram for log of sample of 50 6 8 10 • The median and range are useful summaries of how data are distributed. They should be calculated when the data are not (approximately) normally distributed. 0 2 4 Frequency • The mean and standard deviation are useful summaries of how data are distributed. They should be calculated only when the data are approximately normally distributed. 1 2 3 4 5 range=[0.8,5.5] Now, drawing another random sample of 50 gives : mean=3.0 , SD=0.85 , median=3.0 , range=[1.3,5.0] and again : mean=2.9 , SD=1.00 , median=2.9 , range=[0.4,5.6] and again : mean=3.1 , SD=1.07 , median=3.1 , range=[0.7,4.9] We conclude that for the right skewed data the mean and SD are highly variable, for the normal data the mean and SD, however, provides a very effective summary. The median stays constant for both distributions. 14 Statistical Models When a physical quantity is measured several times we will get different results due to measurement error and biological variation. For example, measuring the height of a subject may yield the following histogram : Estimation in Statistical Models In a statistical model one wishes to learn primarily about the parameters of the model. However, to understand what can be learned about these one must also study the variability present. In the statistical model 0.8 13 0.6 Yi = µ + ǫi i = 1, ..., 200 189 Individual measurement = overall mean + noise If we let the individual measurements be called Yi (the observed data) the overall mean µ (unknown), and the noise ǫ, we have that Y i = µ + ǫi This is a statistical model that describes how the observed measurements arises. The model claims that the individual observations varies around a fixed value (µ), and that the variation is ǫ. A model contains two parts: a systematic part which is of scientific interest and a random variation part which is due to biological and measurement error variation. To complete the specification of the model we also specify how the random variation ǫi varies. We do this by specifying its distribution. It is assumed that ǫi ∼ N (0, σ 2), i.e., it is normal with mean 0 and variance σ2. 15 ȳ = µ̂ = µ + n 1 X ǫi n i=1 The last term is an average of independent noise terms N (0, σ 2) and 2 mathematical arguments give that it is distributed as N (0, σn ). So we have described exactly what is known about µ in µ̂ through finding its distribution (N (µ, σ 2/n)). One way to think about this is that we have a description of how the sample average is varying if we repeat the sampling. The variance of the average is n times smaller than the variance of the individual noise terms. Normal densities 1.5 188 x 1.0 187 normal density 186 0.5 185 What we see is variation around the average height. The variation is due to both measurement error and biological variation. Based on the above histogram it appears reasonable to claim that the variation may be described by a normal distribution. We may phrase this as a statistical model : where ǫi ∼ N (0, σ ) are independent noise terms. We want to know µ and σ. We may estimate these quantities by the sample average and standard deviation. n 1 X Yi ȳ = µ̂ = n i=1 and v u u n 1 X u (Yi − ȳ)2 SD = σ̂ = t n − 1 i=1 Looking at ȳ and using the statistical model we get that 0.0 0.0 0.2 0.4 2 −4 −2 0 2 x 16 4 for log of sample of and Normal Approximation 30 40 Histogram 20 200 0 40 10 Histogram Sperm analysis, cont’d Drawing the best guess at how the population is distributed against the histogram : Frequency Sperm analysis Scientific interest in level of sperm concentration in Danish population. We have representative sample from population. We wish to see if the level in Denmark is equal to what WHO considers the minimum level (20 mill/ml). A sample of 200 Danish men look like this : 2 3 4 5 6 2 3 4 5 6 The log-transformed data appears to be distributed as a normal distribution. A statistical model is now proposed to describe how the population is varying, containing a systematic part (µ) which is the average log(sperm concentration) in the population and a random variation part ǫi, which is independent normal random variation N (0, σ 2) : Yi = µ + ǫi i = 1, ..., 200 We do not know µ and σ. We may estimate these quantities by the sample average and standard deviation. under Null and Observed 0.5 * 20 * dnorm(x + 4, log(20), slx) 30 n 1 X Yi = 3.9 n i=1 Distribution and 0 10 ȳ = µ̂ = We see that the histogram and the normal curve approximate each other well. So the statistical model is validated. Which means that we have a reasonable description of the level of random variation, and a reasonable description of the systematic variation. We wish to investigate if the data is consistent with the null-hypothesis H0 : µ = log(20), if this is not so, we are left with the alternative HA : µ 6= log(20). The meaning of ”consistent with the null-hypothesis” is in statistical terms equivalent to checking if the data could arise when the null-distribution is true. The null-hypothesis claims that the data is distributed around log(20), and if we use the description of the variation found above, the data should arise as a random sample from the left hand curve : 20 1 40 0 10 20 Frequency 30 1 v u u u t n 1 X (Yi − ȳ)2 = 0.95 n − 1 i=1 This means that our best guess is that the population has mean 3.9 and the level of random variation is described by a normal distribution with standard deviation equal to 0.95 SD = σ̂ = 0 2 4 6 8 The right hand curve is the normal approximation to the data. Formally we write Yi = log(20) + ǫi i = 1, ..., 200 ǫ ∼ N (0, 0.952). 17 18 Sperm analysis, cont’d The question now is : how well does this fit with the average we found in our data at 3.9 ? Sperm analysis, The t-test To further summarize how the observed sample average compares to the null-hypothesis we can calculate how many standard deviations it is different from the null-hypothesis : The sample average is distributed as N (µ, σ 2/n), so if H0 is true, the sample average is varying around log(20) with a standard deviation at √ √ σ/ n (which we estimate as σ/ n = 0.95/14 = 0.05). Thus our guess at how the average is varying under the null is N(log(20),(0.05)2). 6 Distribution of Mean under Null (log(20) T = ȳ − log(20) √ = −18 SD/ n which is t-distributed with n − 1 = degrees of freedom (p < 0.0001). We √ define SEM = SD/ n, the standard error of the mean. A t-distribution is varying slightly more than a normal : t−dist f=19 and Normal t−dist f=9 and Normal 0.4 0.4 0.3 0.3 0.3 3 0.2 dnorm(x, 0, 1) 0.2 dnorm(x, 0, 1) 0.2 dnorm(x, 0, 1) 2 0 2 4 6 0.1 0.1 0.1 1 0 8 How well does this fit with the data ?? −4 −2 0 x 2 4 0.0 0.0 x + 4 0.0 dnorm(x + 4, log(20), slx/200^0.5) 4 0.4 5 t−dist f=199 and Normal −4 −2 0 2 x 4 −4 −2 0 2 4 x because we had only a variable guess on the SD of the population. Note that the t-test is on the form observed − expected T = standard errror of observed We now calculate the chance of getting a test-statistic as extreme as or more extreme than the observed one. The chance is computed under the null H0 (the p-value). The smaller this chance is the more evidence against the null. If the p-value is less than 5% we reject the null (at a 5 % level). 19 20 Statistical Models The random variation in a statistical model is described by a distribution. Often a normal distribution. The random variation may consist of several components depending on the context. Different sources may be : • Measurement error. Statistical Models, Summary The recipe when doing statistical analysis : • Scientific hypothesis is formulated. • • Make graphs of data, to get a feel for the data, and the variability. • Statistical model is proposed and validated. • Inter-individual variation. – Systematic variation, contains parameters about which the scientific hypotheses is formulated. • Intra-individual variation. – Random variation described as normal N (0, σ 2). • Variation over time. • Inference about parameters may be drawn in statistical model. The random variation is not the object of interest but we must anyway specify a model for it that appears reasonable to correctly understand how much that can be learned about the systematic part of the variation. 22 21 One-sample Comparison’s, the t-test Consider the 55 ecological farmers and the 141 airline workers : Airline 40 Frequency 20 0 5 0 0 100 200 300 400 0 100 200 eco 300 400 sas We now wish to investigate if the sperm-level is equal to the level 40 mill/ml (found in the literature) for the group of ecological farmers. A statistical model is Yi = µ + ǫi i = 1, ..., 55 where ǫ ∼ N (0, σ 2) are independent noise terms. We know that the data is approximately normal when considered on a log-scale : log−ECO Histogram log−SAS 15 20 15 0 0 5 5 10 Frequency 10 Frequency ȳ − log(40) = 0.51/0.14 = 3.6 SEM which should be looked up in t-distribution with 54 = 55 − 1 degrees of √ freedom, where SEM=SD/ n. We get a p-value at 0.001. Thus, if the null was true and we drew 55 men from the population we would get an average as different or more than the observed average with a chance at 0.001. We conclude that the sperm-level is significantly higher than 40 mill/ml in population of ecological farmers. A 95 % confidence interval for mean-values we can not reject by a 5 % test are : √ √ (µ̂ − 1.96 · SD/ n, µ̂ + 1.96 · SD/ n) (4.2 − 1.96 · 0.14, 4.2 + 1.96 · 0.14) = (3.9, 4.4) This is the range of values for the mean of the sperm-concentration we believe in. 25 Histogram The t-test The one-sample t-test for the hypothesis H0 : µ = log(40) versus HA : µ 6= log(40). The null claims that we see is a sample from a population that varies symmetrically around log(40). T-test for H0 is T = 10 Frequency 15 60 20 80 Organic farmers 2 3 4 log(eco[eco 5 > 6 0 0]) 1 2 3 log(sas[sas 4 > 5 6 0]) and therefore investigate the scientific hypothesis on this scale. Estimate µ and σ by sample average and sample standard deviation n 1 X ȳ = µ̂ = Yi = 4.2 n i=1 and v u u n 1 X t (Yi − ȳ)2 = 0.96 SD = σ̂ = u n − 1 i=1 23 24 Two-sample Comparison’s, the t-test Consider the 55 ecological farmers and the 141 airline workers on a log-scale : Histogram log−SAS 25 Histogram log−ECO 20 15 A Non-parametric One-sample Test, The signed-rank test Non-parametric techniques avoids the assumption of normally distributed residuals, and instead ask questions about the median for the population. Still looking at the ecological farmers. We now take a subset of 10 men: 15 10 Frequency 10 We make a Wilcoxon one-sample test a signed rank test. Subtracting 40 from each of the sperm levels we get -18 3.5 -4 1 15 18 2 3.5 30 5 34 6 40 7 49 8 80 9 160 10 Ordering these after absolute size and assigning them ranks. We check if the sum of the rank’s of the negative values are as big as the ranks of the positive values, as it should be under symmetry. The ranks of the negative numbers are 4.5. We look it up in statistical table. The p-value is p > 0.01 and p < 0.02. Doing the test on all the data gives a p-value at 0.001. 0 H0 : Distribution symmetric around 0 versus HA : Distribution not symmetric. (skewed for example) 0 5 5 and wish to test if they vary symmetrically around 40 mill/ml. We do not specify a detailed statistical model but want to test if Frequency 22 36 55 58 70 74 80 89 120 200 2 3 4 5 6 0 log(eco[eco > 0]) 1 2 3 4 5 6 log(sas[sas > 0]) One may want to know if these two groups really could be varying around the same level, and that the differences we see is due to random variation. We start by proposing a statistical model in which we can answer the question: Yi,j = µi + ǫi,j i = 1, 2, j = 1, ...ni where ǫi,j ∼ N (0, σi2) are independent noise terms. The histograms of the data shows that the model is a good description of the data on log-scale. Estimating the mean and variability in the two populations underlying the samples give that µ1 = 3.9 µ2 = 4.2 σ12 = 1.08 σ22 = 0.90 One may use a normal approximation to compute the p-value, i.e., r compute µ = n(n + 1)/4 and σ = n(n + 1)(2n + 1)/24, and T −µ σ for n > 20. For smaller values of n use a table. Z= 25 26 Two-sample Comparison’s, the t-test To carry out a two-sample t-test we first need to check if the variability is the same in the two groups. We test if H0 : σ1 = σ2 versus HA : σ1 6= σ2. And use the following test-statistic : Non-parametric Two-sample Comparison’s, The rank test The non-parametric rank test is also called the Wilcoxon-Mann-Whitney test. Consider two groups of data as before. We now wish to test if the distribution of the two population could be equal, or if this must be rejected by a test. The statistical model : • : Yi,j ∼ arbitrary distribution Fi (·). • : All data points are independent. max(σ12, σ22) min(σ12, σ22) 1.08 = = 1.27 0.90 F = which we should look up in F − distribution with (140, 54) degrees of freedom (p=0.32). So we accept hypothesis. Now we can calculate a combined estimate of the variability (n1 − 1)σ12 + (n2 − 1)σ22 (n1 − 1) + (n2 − 1) 54 · 0.902 + 140 · 1.082 = = 1.02 55 − 1 + 141 − 1 SD 2 = With the combined variability estimate SD we can proceed to the twosample T-test for H0 : µ1 = µ2 versus HA : µ1 6= µ2 T = y¯1 − y¯2 = 2.82 SD (1/n1) + (1/n2) r which we look up in t-distribution with n1 + n2 − 2 = f1 + f2 degrees of freedom. (p=0.006). • We conclude that the ecological farmers have a significantly higher sperm-level than the airline workers. A 95 % confidence interval for the difference in means of the two groups are given by : (y¯1 − y¯2 − 1.96 · SED, y¯1 − y¯2 + 1.96 · SED) = (0.3 − 2 · 0.1, 0.3 + 2 · 0.1) r where SED = SD ∗ ( (1/n1) + (1/n2)). 27 In this non-parametric model we wish to test if : H0 : Distributions are the same versus HA : Distributions are not the same. We calculate a test-statistic as follows: • Pool all data and assign ranks. • Sum ranks of smallest group. • Look sum of ranks up in statistical table to get p-value. Sum of ranks, T, for ecological farmers is 6342 (total sum of ranks is 19306, and 19306 * (55/196) = 5405) which result in p-value at 0.0096 (computer program). One may use a normal approximation to compute the p-value, i.e., r compute µ = n1(n1 + n2 + 1)/2 (5390) and σ = n2µ/6 (356), and T −µ σ for n1, n2 > 10. For smaller values, use a table. Z= 28 Paired Comparison’s When data is paired the two measurements often are not independent: • Measuring right- and left bicep. • Growth before and after treatment. • Height of men of women when sampled as couples. With only two correlated measurements, the data may anyway be analysed by simple techniques. A correct analysis is obtained by making one-sample analysis on the differences. The differences between the before and after measurements are namely independent among subjects. Therefore one should simply test if the differences are varying around 0, by either a t-test or a signedrank-test. When investigating the effect of some drug that prevents sun-burn, say, we could apply the sun-lotion to one arm and placebo to the other. The difference between the arms may be ascribed to the lotion. The difference is a measure that is corrected for inter-individual variation, which may be large. Summary • Make graphs of data. One-sample test: • When the variation is approximately normal the t-test may be used to test a hypothesis about the mean of the underlying population. The p-value provided is only valid if the variation is approximately normal. A nice summary of data is provided by the confidence interval of the mean. • When data is not normally distributed and interest is concentrated on inference rather than estimates the signed-rank-test may be used. This test is always valid. No confidence intervals may be given. Right skewed data may be transformed to approximate normality by transformations √ like x, x1/3, log. Two-sample test: • Two groups of data may be compared by the t-test when the variation is approximately normal and the variance of the residual variation is equal in the two groups. A nice summary of difference between the groups are given by the confidence interval for the difference between the means. • When data is not normally distributed and interest is concentrated on inference rather than estimates the rank-test may be used. This test is always valid. No confidence intervals may be given. • Paired data is handled by sample techniques on the differences between the pairs. 29 30 Statistical Analysis using Analyst (SAS) Analyst is a windows based application in the SAS statistical software. SAS is activated by clicking : start → statistik → SAS in the lower lefthand corner. Analyst is activated after solutions → analysis → Analyst Commands will be presented as we need them for the various analyses, and remember that the focus is on the statistical analyses rather than how one do this and that. We consider data on sperm concentration (mill/ml) on two groups of people in a study. One group are members of an association that promotes the development of organic agriculture (n=55), and another group of workers from a major Scandinavian airline carrier (n=141). now type a new name e.g. oeko12 if you are in from of machine 12. The data-set contains the following variables : obs observation number. abstime length of abstinence in days. age age of subject. s1e2 group indicator. conc sperm concentration (mill/ml). volume volume of sperm sample (ml). The data is loaded file → open... from n:\human\oeko that is a SAS data-set. Doing this the data will appear in the data table. It consists of a record for each subject with the variables described above. To make your own new variables when you work with the data you must create your own version of the data. You do this by saving your own version of the data under a new name : File → Save... → 31 32 Data Manipulations A little bit of data manipulation is needed. New transformed variables are constructed by setting the data frame in edit mode edit → mode → edit and then data → transform → compute... now type new variable name (e.g. conc3) and an expression that defines the new variable in the box below the equality-sign (e.g. conc**.3333). Now, a new variable called conc3 that is equal to concentrations on cube-root–scale is defined and appears in the data table. √ Data Manipulations To group a continuous variable according to its value and to define a classification variable based on it : data → transform → recode ranges... in the recode dialog give column name (volume) and name of the new grouped version (gvol) and click ok. Now in the next window give the bounds 0,3; 3,4, and 4,15 for the first three groups and name them (1,2,3) in the rightmost column, click ok. To delete variable highlite the column in the data-table : edit → delete Alternatively, one may take on of the standard transformations like conc after highlighting the column one wishes to transform by data → transform → √ To make a variable that can be used for the one-sample test (e.g. ld40=lconc-log(40)) data → transform → compute... now type new variable name (ld40) and the expression that defines the new variable in the box below the equality sign log(conc)-log(40). To construct a subset of the data, e.g., the subset of ecological farmers for an specific analysis for this group : data → filter → subset data... in the subset dialog you can apply a Where clause to the data (click s1e2 and eq and constant value followed by 1 to select s1e2=1 the Airline workers). 33 34 Histograms To make a histogram of concentration ( conc ) graphs → histogram... select conc as the analysis variable and s1e2 as the class variable (the classification variable). If the class variable is omitted no-classification variable will used. Now, clicking ok does the job. Simple descriptive Statistics To compute mean, standard deviations, variances, medians and percentiles as well as the range statistics → descriptive → distributions... select conc as the analysis variable and s1e2 as the class variable (the classification variable). Now, clicking ok does the job. Output Airline 80 Organic farmers 40 Frequency 0 0 5 20 10 Frequency 15 60 20 ----------------------------- S1E2=1 -----------------------------Univariate Procedure Variable=CONC Moments N 141 Sum Wgts 141 Mean 69.16461 Sum 9752.21 Std Dev 70.17659 Variance 4924.753 Skewness 2.172157 Kurtosis 5.780222 USS 1363973 CSS 689465.5 CV 101.4631 Std Mean 5.909935 T:Mean=0 11.70311 Pr>|T| 0.0001 Num ^= 0 139 Num > 0 139 M(Sign) 69.5 Pr>=|M| 0.0001 Sgn Rank 4865 Pr>=|S| 0.0001 0 100 200 300 400 0 eco 100 200 300 400 Quantiles(Def=5) sas To examine the normality of a variable one may draw the histogram for a normal distribution on the same plot. To do this click fit in the distribution-dialog and and select normal and ok in the fit-dialog before clicking ok on the distribution-dialog. 100% 75% 50% 25% 0% Range Q3-Q1 Mode Max Q3 Med Q1 Min 402 91 48 23 0 99% 95% 90% 10% 5% 1% 358 209 158 12 3.3 0 402 68 12 Extremes Lowest Obs Highest Obs 0( 40) 233( 92) 0( 1) 284( 102) 0.75( 67) 308( 32) 1.88( 60) 358( 104) 2.3( 132) 402( 69) ----------------------------- S1E2=2 -----------------------------Univariate Procedure 35 36 Variable=CONC Moments 55 Sum Wgts 99.04727 Sum 86.39382 Variance 1.339362 Kurtosis 942620.1 CSS 87.22483 Std Mean 8.502394 Pr>|T| 54 Num > 0 27 Pr>=|M| 742.5 Pr>=|S| N Mean Std Dev Skewness USS CV T:Mean=0 Num ^= 0 M(Sign) Sgn Rank 55 5447.6 7463.891 1.118476 403050.1 11.64934 0.0001 54 0.0001 0.0001 ----------------------------- S1E2=1 -----------------------------Univariate Procedure Variable=DL40 Moments Quantiles(Def=5) 100% 75% 50% 25% 0% Max Q3 Med Q1 Min 354 138 69 33 0 Range Q3-Q1 Mode One-sample T-test and Signed Rank Test We wish to examine if the hypothesis that the sperm level varies around 40 mill/ml can be statistically rejected or validated. To make a one-sample t-test first transform to log-scale to obtain approximate normality and then compute a new variable dl40=lconc-log(40) (see above). Now, statistics → descriptive → distributions... selecting the variable dl40 and with class equal to s1e2 does the job. Output: 99% 95% 90% 10% 5% 1% 354 297 259 15 9.1 0 354 105 69 N Mean Std Dev Skewness USS CV T:Mean=0 Num ^= 0 M(Sign) Sgn Rank 139 0.091883 1.080798 -0.79219 162.3748 1176.271 1.002305 139 9.5 816 Sum Wgts Sum Variance Kurtosis CSS Std Mean Pr>|T| Num > 0 Pr>=|M| Pr>=|S| 139 12.7718 1.168125 1.421361 161.2013 0.091672 0.3180 79 0.1265 0.0862 Extremes Quantiles(Def=5) Lowest 0( 5.5( 9.1( 11( 14( Obs 40) 15) 35) 42) 10) Highest 264( 264( 297( 322( 354( Obs 32) 33) 47) 14) 51) 100% 75% 50% 25% 0% Max Q3 Med Q1 Min Range Q3-Q1 Mode 2.307573 0.832909 0.182322 -0.51083 -3.97656 99% 95% 90% 10% 5% 1% 2.191654 1.704748 1.373716 -1.20397 -1.63476 -3.05761 6.284134 1.343735 -1.20397 Extremes Lowest -3.97656( -3.05761( -2.85597( -2.69563( -2.56395( Obs 67) 60) 132) 111) 49) Highest 1.762159( 1.960095( 2.04122( 2.191654( 2.307573( 37 Moments 54 Sum Wgts 0.541879 Sum 0.958596 Variance -0.50364 Kurtosis 64.55813 CSS 176.9023 Std Mean 4.153971 Pr>|T| 54 Num > 0 14 Pr>=|M| 428.5 Pr>=|S| 54 29.26144 0.918905 -0.00368 48.70198 0.130448 0.0001 41 0.0002 0.0001 Quantiles(Def=5) 100% 75% 50% 25% 0% Max Q3 Med Q1 Min 2.180417 1.238374 0.545227 0.09531 -1.98413 Range Q3-Q1 Mode 99% 95% 90% 10% 5% 1% 2.180417 2.004853 1.867949 -0.85567 -1.29098 -1.98413 4.164549 1.143064 0.545227 Obs Highest 15) 1.88707( 35) 1.88707( 42) 2.004853( 10) 2.085672( 29) 2.180417( Missing Value Count % Count/Nobs One-sample T-test Alternatively one may use a special menu that has been designed especially for the one-sample t-test statistics → hypothesis tests → One-sample t-test... selecting the variable lconc and entering the mean we wish to test as 4. Note that the t-test should be carried out only the group of ecological farmers, say, and that the active data-set therefore should be only this group. To make the test it is necessary to construct a new data set that consists of the group of interest as done in the data manipulation section above. Output: One Sample T Test for a Mean Sample Statistics for LCONC N Mean Std. Dev. Std. Error ------------------------------------------------193 3.91 1.07 0.08 Hypothesis Test Null hypothesis: Alternative: Mean of LCONC = 4 Mean of LCONC ^= 4 t Statistic Df Prob > t ---------------------------------1.217 192 0.2249 Extremes Lowest -1.98413( -1.48061( -1.29098( -1.04982( -0.98083( 92) 102) 32) 104) 69) 38 Missing Value . Count 2 % Count/Nobs 1.42 ----------------------------- S1E2=2 -----------------------------Univariate Procedure Variable=DL40 N Mean Std Dev Skewness USS CV T:Mean=0 Num ^= 0 M(Sign) Sgn Rank Obs Obs 32) 33) 47) 14) 51) To make the t-test of the two groups, you can specify that you want it done for the two groups under the variables button, by given s1e2 as the by variable. . 1 1.82 39 40 Two-sample T-test for Means (un-paired data) To compare the concentrations for the two groups statistics → hypothesis tests → Two-sample t-test for means... selecting the variable lconc and the group variable s1e2. Output: Two Sample T Test for the Means of LCONC within S1E2 Sample Statistics Group N Mean Std. Dev. Std. Error -------------------------------------------------1 139 3.780763 1.0808 0.0917 2 54 4.230758 0.9586 0.1304 Two-sample T-test for Variances (un-paired data) To compare the concentrations for the two groups statistics → hypothesis tests → Two-sample t-test for variances... selecting the variable lconc and the group variable s1e2. Output: Two Sample Test for Variances of LCONC within S1E2 Sample Statistics S1E2 Group N Mean Std. Dev. Variance -------------------------------------------------1 139 3.7808 1.0808 1.1681 2 54 4.2308 0.9586 0.9189 Hypothesis Test Hypothesis Test Null hypothesis: Alternative: Null hypothesis: Alternative: Mean 1 - Mean 2 = 0 Mean 1 - Mean 2 ^= 0 If Variances Are t statistic Df Pr > t ---------------------------------------------------Equal -2.677 191 0.0081 Not Equal -2.822 108.14 0.0057 Variance 1 / Variance 2 = 1 Variance 1 / Variance 2 ^= 1 - Degrees of Freedom F Numer. Denom. Pr > F ---------------------------------------------1.27 138 53 0.3203 It is useful to supplement the analysis with some plots. Try for example the plots button, and select one of the plots. The conclusions are based on an assumption of equal variances, and this should be validated. The output may indicate that this is the case, but if in doubt one can carry out a test that shows have serious the deviation from equal variances are. 41 42 Two-Sample Signed Rank Test The two-sample signed rank test can more generally by considered as a special case of the Kruskal-Wallis test that test if k groups have the same distribution. To carry out the two-sample signed rank test : statistics → ANOVA → non-parametric one-way ANOVA... selecting the variable conc and the group variable s1e2. Output: Exercise-I Rather than considering the concentration we shall now consider the volume of each sperm sample as the parameter of interest. We wish to compare the ecological farmers and the airline workers. A volume of 3 ml is considered normal. Investigate further if the two groups are normal in this respect. Wilcoxon Scores (Rank Sums) for Variable CONC Classified by Variable S1E2 S1E2 1 2 N 141 55 Sum of Scores Expected Under H0 Std Dev Under H0 Mean Score 12964.0 13888.5000 356.782425 91.943262 6342.0 5417.5000 356.782425 115.309091 Average Scores Were Used for Ties 3) Without doing any computer work make a strategy for how such an analyses can and should be carried out. What descriptive plots and statistics are needed ? What hypothesis are formulated and tested ? How will you validate the necessary assumptions for the suggested analysis ? 4) Do the analyses, make the plots and so on. Remember to interpret the results according to the subject matter. Wilcoxon 2-Sample Test (Normal Approximation) (with Continuity Correction of .5) S = 6342.00 Z = 2.58981 Prob > |Z| = 0.0096 T-Test Approx. Significance = 0.0103 Kruskal-Wallis Test (Chi-Square Approximation) CHISQ = 6.7144 DF = 1 Prob > CHISQ = 0.0096 43 44