* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download 3D anatomical modelling of the human cardiac conduction system
Heart failure wikipedia , lookup
Cardiac contractility modulation wikipedia , lookup
Rheumatic fever wikipedia , lookup
Quantium Medical Cardiac Output wikipedia , lookup
Aortic stenosis wikipedia , lookup
Mitral insufficiency wikipedia , lookup
Lutembacher's syndrome wikipedia , lookup
Myocardial infarction wikipedia , lookup
Artificial heart valve wikipedia , lookup
Cardiac surgery wikipedia , lookup
Dextro-Transposition of the great arteries wikipedia , lookup
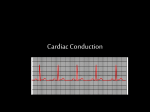
Electrocardiography wikipedia , lookup
Arrhythmogenic right ventricular dysplasia wikipedia , lookup
3D anatomical modelling of the human cardiac conduction system A thesis submitted to the University of Manchester for the degree of Master of Philosophy in the Faculty of Medical and Human Sciences. 2014 Andrew Atkinson School of Medicine Word Count:23725 CONTENTS LIST OF FIGURES ..................................................................................................... 5 LIST OF MOVIES ....................................................................................................... 7 LIST OF TABLES ....................................................................................................... 8 ABSTRACT ................................................................................................................. 9 DECLARATION ....................................................................................................... 10 COPYRIGHT STATEMENT .................................................................................... 11 ABBREVIATIONS ................................................................................................... 12 ACKNOWLEDGEMENTS ....................................................................................... 15 CHAPTER 1) INTRODUCTION ............................................................................. 16 1.1) The cardiac conduction system .................................................................... 16 1.2) Key Components of CCS ............................................................................ 18 1.2.1) Sinus node ......................................................................................................... 18 1.2.2) Novel paranodal area ................................................................................... 25 1.2.3) Internodal pathways..................................................................................... 31 1.2.4) Atrioventricular node ................................................................................... 32 1.2.5) Ventricular conduction system................................................................. 37 1.3) Analysis of existing cardiac models ............................................................ 39 CHAPTER 2) GENERAL METHODS ..................................................................... 45 2.1) Human sample details and ethical approval ................................................ 45 2.2) General principles ........................................................................................ 45 2.2.1) Immunohistochemistry ............................................................................... 45 2.2.2) Laser Scanning Confocal Microscopy ..................................................... 54 2.2.4) Histology............................................................................................................ 61 2.2.5) Micro-CT ............................................................................................................ 63 CHAPTER 3) 3D ANATOMICAL RECONSTRUCTION OF THE COMPLETE HUMAN SINUS NODE AND PARANODAL AREA ............................................. 65 3.1) Introduction ................................................................................................. 65 3.1.1) Aim ........................................................................................................... 65 3.2) Materials and methods ................................................................................. 65 3.2.1) Sample preparation....................................................................................... 65 3.2.2) X-Ray Computed Tomography.................................................................. 67 3.2.3) Tissue sectioning ............................................................................................ 67 3.2.4) Histology............................................................................................................ 67 2 3.2.5) Immunohistochemistry ............................................................................... 68 3.2.6) 3D anatomical reconstruction .................................................................. 68 3.3) Results ......................................................................................................... 71 3.4) Discussion .................................................................................................... 76 3.5) Limitations and future work ........................................................................ 78 CHAPTER 4) 3D ANATOMICAL RECONSTRUCTION OF THE COMPLETE CARDIAC CONDUCTION SYSTEM OF THE HUMAN HEART ........................ 79 4.1) Introduction ................................................................................................. 79 4.2) Materials and methods ................................................................................. 79 4.2.1) Whole human heart....................................................................................... 79 4.2.2) X-Ray Computed Tomography.................................................................. 80 4.2.3) Tissue preparation ........................................................................................ 80 4.2.4) 3D anatomical reconstruction .................................................................. 83 4.3) Results ......................................................................................................... 83 4.4) Discussion .................................................................................................... 90 4.4.1) Proximity of aortic valve to components .............................................. 90 4.4.2) Uses of the anatomical model ................................................................... 95 4.5) Limitations and future work ........................................................................ 96 CHAPTER 5) INVESTIGATING THE EXPRESSION OF THE LARGE CONDUCTANCE KCA1.1 CHANNEL IN THE HUMAN SINUS NODE .............. 98 5.1) Introduction ................................................................................................. 98 5.2) Materials and methods ............................................................................... 100 5.2.1) Molecular investigation ............................................................................ 100 5.2.2) Protein investigation ................................................................................. 104 5.3) Results ....................................................................................................... 104 5.3.1) qPCR ................................................................................................................. 104 5.3.2) KCa protein investigation .......................................................................... 105 5.4) Discussion .................................................................................................. 111 5.5) Limitations ................................................................................................. 114 CHAPTER 6) GENERAL DISCUSSION ............................................................... 115 REFERENCES......................................................................................................... 117 PUBLICATIONS ..................................................................................................... 125 Full papers and book chapters ........................................................................... 125 Abstracts ........................................................................................................... 125 3 APPENDIX .............................................................................................................. 127 4 LIST OF FIGURES Figure 1.1. Anatomical location of sinus node. ........................................................ 19 Figure 1.2. Histology and immunohistochemistry for Cx43 of human sinus node and surrounding tissue. ..................................................................................................... 21 Figure 1.3. Histological sections through the crista terminalis at various levels. ..... 22 Figure 1.4. 3D anatomical model of the human sinus node. ..................................... 24 Figure 1.5. Sinus node activation sites and pathways. .............................................. 26 Figure 1.6. Optical mapping of sinus node. .............................................................. 28 Figure 1.7. Location of the atrioventricular node. .................................................... 33 Figure 1.8. Understanding pathways of the atrioventricular node. ........................... 36 Figure 1.9. Anatomy of the human His-Purkinje system. ......................................... 38 Figure 1.10. Models of human cardiac conduction system and activation. .............. 40 Figure 1.11. Comparison between histology and OCT. ............................................ 44 Figure 2.1. Basic antibody structure. ........................................................................ 48 Figure 2.2. Major components and principle of a confocal microscope. .................. 55 Figure 2.3. Principle of SYBR green PCR quantitation............................................ 58 Figure 2.4. qPCR data analysis. ................................................................................ 59 Figure 2.5. Principle of computed tomography. ....................................................... 64 Figure 3.1. Sinus node sample. ................................................................................. 66 Figure 3.2. Histology of sinus node sample. ............................................................. 69 Figure 3.3. Sinus node reconstruction workflow. ..................................................... 70 Figure 3.4. Sinus node anatomical reconstruction. ................................................... 72 Figure 3.5. Sinus node extensions. ............................................................................ 73 Figure 4.1. Histology on whole human heart sections. ............................................. 81 Figure 4.2. Histology on serial whole human heart sections. ................................... 82 Figure 4.3. Whole heart anatomical reconstruction. ................................................. 84 Figure 4.4. Anterior view of 3D anatomical reconstruction of human CCS. ........... 85 Figure 4.5. Medial view of 3D anatomical reconstruction of human CCS. .............. 86 Figure 4.6. Posterior view of 3D anatomical reconstruction of human CCS. ........... 87 Figure 4.7. CCS reconstruction with atria removed. ................................................. 88 Figure 4.8. Coronal slice through 3D reconstruction at level of aortic valve. .......... 91 Figure 4.9. 12-lead ECG recording from patient undergoing TAVI procedure........ 94 5 Figure 5.1. The effect of paxilline and iberiotoxin on heart rate in the isolated rat heart. ........................................................................................................................... 99 Figure 5.2. mRNA expression in the SN, PN and AM. .......................................... 107 Figure 5.3. Localisation of KCa1.1 protein............................................................. 108 Figure 5.4. APC012 KCa1.1 antibody concentration optimisation. ....................... 109 Figure 5.5. APC151 KCa1.1 antibody concentration optimisation. ....................... 110 Figure 5.6. KCa1.1 expression in SN, PN and AM quantification........................... 112 Figure 5.7. Pacemaking mechanisms in the sinus node .......................................... 113 Figure A1. Sample of outlined images used for sinus node reconstruction. ........... 128 Figure A2. Comparison of whole heart histology and CT images. ......................... 129 Figure A3. Computer simulation created using the whole heart 3D anatomical reconstruction ........................................................................................................... 130 Figure A4. KCa1.1 Localisation ............................................................................... 131 6 LIST OF MOVIES Movie 3.1. 3D anatomical reconstruction of SN and paranodal area. Movie 4.1. 3D anatomical reconstruction of the human heart. 7 LIST OF TABLES Table 2.1. Heart details ............................................................................................. 46 Table 2.2. List of primary antibodies used ................................................................ 51 Table 2.3. Excitation and emission wavelengths of secondary antibody dyes ......... 52 Table 2.4. List of secondary antibodies used ............................................................ 53 Table 3.1. Sinus node reconstruction statistics. ........................................................ 74 Table 3.2. Comparison of sinus node sizes. .............................................................. 75 Table 4.1. Whole heart model statistics. ................................................................... 89 Table 5.1. qPCR primers used................................................................................. 103 8 ABSTRACT The University of Manchester Andrew James Atkinson Submission for the degree of Master of Philosophy 3D anatomical modelling of the human cardiac conduction system June 2014 Background. The cardiac conduction system is responsible for the initiation and propagation of action potentials in the heart. The sinus node is the primary pacemaker in the heart. Action potentials then pass through the atrial myocardium to the atrioventricular node, into the His bundle and finally the Purkinje fibre network. A detailed understanding of the anatomy of the cardiac conduction system is important in understanding its functioning and potential role in arrhythmogenesis. The creation of 3D models is an increasingly important field as they are powerful tools and have many potentially valuable educational, research and clinical uses such as teaching complex anatomy can and in computer modelling studies of disease. Aims. In this study the aim was to create 3D anatomical computer models of the human sinus node and complete paranodal area, and also create a reconstruction of the major components of the cardiac conduction system of the human heart from an intact heart. Methods. A formalin fixed human heart and a right atrial sample containing the intercaval region were CT scanned. The samples were then frozen and serially sectioned. Masson’s trichrome histology and immunohistochemistry using antibodies against Cx43, HCN4 and KCa1.1 were used to identify the major components of the cardiac conduction system. Avizo software was used to segment structures and create 3D anatomical reconstructions. 3D measurements of structures made. Results. 1) A 3D anatomical reconstruction of the sinus node and paranodal was successfully created. The sinus node is a crescent shaped structure 1.6 cm in length. The paranodal area is ellipsoid in shape and 2 cm in length. The two structures are in close proximity but there were no connections found between them. 2) A 3D anatomical reconstruction of the human heart and major components of the cardiac conduction system was successfully created from tissue sections of the intact heart. The His bundle is the closest structure to the non-coronary cusp of the aortic valve. 3) KCa1.1 appears to be more highly expressed in the sinus node than atrial muscle at both the mRNA and protein level. Discussion. This study demonstrates that it is possible to create a 3D anatomical reconstruction of the human heart based on serial sections of the entire human heart. This 3D computer anatomical model has many potential clinical and educational uses. The model can be used in computer modelling studies into cardiac pacemaking and arrhythmia and particularly in relation to aortic valve replacement, this model may be useful in helping improve the design of the implants in order for them to have less of an impact on the cardiac conduction system. The KCa1.1 channel may be a potential target in the treatment of arrhythmia. 9 DECLARATION No portion of the work referred to in the thesis has been submitted in support of an application for another degree or qualification of this or any other university or other institute of learning 10 COPYRIGHT STATEMENT The following four notes on copyright and the ownership of intellectual property rights must be included as written below: i. The author of this thesis (including any appendices and/or schedules to this thesis) owns certain copyright or related rights in it (the “Copyright”) and s/he has given The University of Manchester certain rights to use such Copyright, including for administrative purposes. ii. Copies of this thesis, either in full or in extracts and whether in hard or electronic copy, may be made only in accordance with the Copyright, Designs and Patents Act 1988 (as amended) and regulations issued under it or, where appropriate, in accordance with licensing agreements which the University has from time to time. This page must form part of any such copies made. iii. The ownership of certain Copyright, patents, designs, trademarks and other intellectual property (the “Intellectual Property”) and any reproductions of copyright works in the thesis, for example graphs and tables (“Reproductions”), which may be described in this thesis, may not be owned by the author and may be owned by third parties. Such Intellectual Property and Reproductions cannot and must not be made available for use without the prior written permission of the owner(s) of the relevant Intellectual Property and/or Reproductions. iv. Further information on the conditions under which disclosure, publication and commercialisation of this thesis, the Copyright and any Intellectual Property and/or Reproductions described in it may take place is available in the University IP Policy (see http://www.campus.manchester.ac.uk/medialibrary/policies/intellectualproperty.pdf), in any relevant Thesis restriction declarations deposited in the University Library, The University Library’s regulations (see http://www.manchester.ac.uk/library/aboutus/regulations) and in The University’s policy on presentation of Theses. 11 ABBREVIATIONS ABC avidin and biotinylated horseradish peroxidase macromolecular complex ANP atrial natriuretic peptide AVN atrioventricular node AVNRT atrioventricular nodal re-entrant tachycardia BK big potassium BSA bovine serum albumin Cav voltage-gated Ca2+ channel CCS cardiac conduction system cDNA complementary DNA (cDNA CT computed tomography Ct threshold cycle Cx40 connexin 40 Cx43 connexin 43 DAB diaminobenzidine DNA deoxyribonucleic acid dNTPs deoxynucleotide triphosphate molecules (dNTPs DT-MRI diffusion tensor magnetic resonance imaging ECG electrocardiogram Eff efficiency F fluorescence Fab fragment, antigen binding region 12 Fc fragment, crystallisable region H&E hematoxylin and eosin HCN hyperpolarization activated cyclic nucleotide-gated potassium channel HeNe helium-neon If funny current IgG immunoglobulin G IgM immunoglobulin M KCa1.1 calcium-activated potassium channel Kir inwardly-rectifying K+ channel Kv4.2 voltage-gated K+ channel MRI magnetic resonance imaging mRNA messenger ribonucleic acid NCX sodium-Calcium exchanger OCT optical coherence tomography PBS phosphate buffered saline PCR polymerase chain reaction PMT photomultiplier qPCR quantitative polymerase chain reaction RNA ribonucleic acid RYR ryanodine receptor SERCA sarcoplasmic reticulum Ca2+-ATPase SK2 small conductance calcium-activated K+ channel 13 SN sinus node TASK tandem-pore acid-sensitive K+ channel TAVI/TAVR transcatheter aortic valve implantation/replacement TBX3 T-Box transcription factor 3 14 ACKNOWLEDGEMENTS I would firstly like to thank my supervisor Dr Halina Dobrzynski for her enthusiastic support and guidance throughout this project. I would also like to thank Professor Mark Boyett and Professor Bob Anderson for their contributions to this work. I am grateful to all of my colleagues at the University of Manchester for their support and advice whilst I was undertaking this project. I am particularly grateful to Miss Sandy Chu for her assistance. Finally I would like to thank my family for their never ending support and encouragement. 15 CHAPTER 1) INTRODUCTION 1.1) The cardiac conduction system The cardiac conduction system (CCS) is responsible for the initiation and propagation of action potentials within the heart. The major components of the CCS are the sinus node (SN), atrioventricular node (AVN), His Bundle, left and right bundle branches and the Purkinje fibres. The earliest recorded studies of the CCS were carried out in the late 19th century, when it was first shown that cardiac impulse conduction was myogenic and did not involve nerves (Stannius, 1852). It was the ventricular components of the CCS that were first to be identified and the work of Tawara in 1906 showed that the AVN, His bundle, bundle branches and Purkinje fibres were in continuity with each other forming a direct myocardial connection between the atria and ventricles (Tawara, 2000). It was not until 1907 that the SN was discovered by Keith and Flack via histological analysis of serial sections through the junctional area between the superior vena cava and the right atrium (Keith and Flack, 1907). It had been observed for some time prior to their work that in hearts that had been removed from cadavers the muscle in this area was the last to stop contracting and that the sinus venosus was site of impulse generation in the heart (Gaskell, 1900; Silverman and Upshaw, 2002). Keith and Flack’s observations were of a “peculiar musculature surrounding artery at the sino-auricular junction”. They discovered that the myocytes in this site were packed within connective tissue and showed similarities to the earlier discovered AVN. It was based on these early histological findings in the SN and AVN that in 1910 guidelines were set down for the criteria to determine whether tissues were to be considered responsible for cardiac conduction. The criteria stated that the tissue should be histologically discrete from adjacent working myocardium, serially traceable from section to section and insulated from the adjacent working myocardium by a sheath of fibrous tissue (Aschoff, 1910; Monckeberg, 1910). The guidelines for the characterisation of the CCS proved adequate to enable the anatomical location of the major components to be determined using histological staining on serial sections through hearts. Using these techniques numerous studies 16 produced detailed representations on the human CCS (Truex et al., 1967; Tawara, 2000; Sanchez-Quintana et al., 2005). However advances in techniques such as immunohistochemistry and advanced medical imaging systems have enabled further analysis on the anatomy of the CCS, including in 3D, and have demonstrated that tissues that do not meet all of the criteria set down are likely to be involved in conduction in the heart. Hyperpolarization activated cyclic nucleotide-gated potassium channel 4 (HCN4) is a protein that is linked to pacemaking in cells as it contributes to the pacemaker current, If, that is involved in spontaneous diastolic depolarization (Shi et al., 1999). Connexin43 (Cx43) is a gap junctional protein and is associated with the working myocardium as it provides strong coupling of cells and possess relatively high conductance properties allowing rapid signal transduction to occur (Jongsma and Wilders, 2000). The SN and AVN show positive immunohistochemical staining for HCN4 and negative staining for Cx43 (Anderson et al., 2009). Immunohistochemical studies using Cx43 and HCN4 antibodies have largely corroborated the earlier histological investigations, however, they have also revealed the existence of additional areas that share the protein expression characteristics of the SN and AVN within the heart that could potentially play a role in pacemaking. Areas such as the atrioventricular ring tissues, located around the atrioventricular valve annuli, in numerous species are HCN4 positive and Cx43 negative (Yanni et al., 2009) and are the target of recent investigations to ascertain their properties and potential functional significance. Quantitative PCR has also expanded the knowledge of the CCS by allowing the genes expressed in the SN and AVN to be identified in many species including the human (Gaborit et al., 2007; Chandler et al., 2009; Greener et al., 2011). Once these tissues were profiled it has been possible to compare the expression found in other tissues such as the atrioventricular rings and further assess whether they should be considered as cardiac conduction tissues (Atkinson et al., 2013). Studies into the development of the heart have also examined the formation of the tissues involved in cardiac conduction. These have identified many transcription factors, such as TBX3, that are critical for the gene expression pattern of the CCS. Tracing the TBX3 expressing tissues through development has further increased the potential number of tissues related to the CCS. These regions include the right venous valve and the AV rings (Sizarov et al., 2011). 17 The structure and function of the CCS has been shown to alter significantly with cardiac disease (Demoulin and Kulbertus, 1978). It was noted by Keith and Flack that diseased hearts they studied appeared to show an increase in fibrosis in the SN and AVN (Keith and Flack, 1907), and understanding these changes is important in the development of better treatments. 1.2) Key Components of CCS 1.2.1) Sinus node The SN is the pacemaker of the heart, initiating cardiac impulses. On inspection of the intact heart the SN is located in the intercaval region of the right atrium, extending from the junction of the superior vena cava and the crest of the right atrial appendage to the inferior vena cava (Fig. 1.1A). Visualising the endocardial surface of the right atrium the SN lies along the superior portion of the crista terminalis, the muscular bundle between the striated atrial appendage and atrial chamber myocardium (Fig. 1.1B). Sectioning of the right atrium perpendicular to the crista terminalis and histological staining, clearly shows a discrete fibrous structure containing small groups of cells situated between the myocardium of the crista terminalis and the superior vena cava (Fig. 1.2A). In the human the SN is commonly punctured by the large sinus node artery (Fig. 1.2A). Serial sectioning shows how the morphology of the node changes as it is traced along the crista terminalis (Fig. 1.3). The extent of the SN has been a source of great intrigue and debate. The SN is classically separated into 3 portions, with the narrower head and tail at the superior and inferior extremities respectively and the wider body in-between (Sanchez-Quintana et al., 2005) (Fig. 1.3). The body forms the majority of the SN (~70%) and is at least twice the height of the head and tail (Sanchez-Quintana et al., 2005). By tracing the node through the serial sections it was possible to create an image of the extent of the SN. Several studies show the node to be an extensive spindle shaped 18 Figure 1.1. Anatomical location of sinus node. 19 Figure 1.1. Anatomical location of sinus node. A. Gross anatomy of human heart, showing approximate location of sinus node. B. Left. Panoramic view of posterolateral wall of the right atrium. Arrowheads indicate sulcus terminalis, junction between the sinus node and the sinus venosus. Right. Endocardial aspect of the same tissue. CT: crista terminalis; FO: foramen ovale; IVC: inferior vena cava; PM: pectinate muscle; RA: right atrium; RAA: right atrial appendage; RSPV: right superior pulmonary vein; SV: sinus venosus; SVC: superior vena cava. Matsuyama et al. (2004). 20 From Figure 1.2. Histology and immunohistochemistry for Cx43 of human sinus node and surrounding tissue. A. Distinctive histological staining of sinus node and diffuse paranodal area in close proximity. Absence of Cx43 expression in sinus node is shown. B. High magnification images of sinus node, paranodal area and right atrium. Loosely packed cells in paranodal area distinctive from sinus node and atrium. Intermediate Cx43 expression seen in paranodal area. From Chandler et al. (2011). 21 Figure 1.3. Histological sections through the crista terminalis at various levels. Sinus node shown in yellow. High magnification images shown in right panel. CT: crista terminalis; IVC: inferior vena cava; SAN: sinus node; SEP: septum; SV: sinus venosus; SVC: superior vena cava. From Matsuyama et al. (2004). 22 structure lying in closer proximity to the epicardium than endocardium, with the head and tail projecting towards the endocardium (Sanchez-Quintana et al., 2005). The total length of the SN has shown some considerable variation between studies. For example Sanchez Quintana et al. (2005) found the SN to average 13.5 mm in length across 47 hearts, with a maximum length of 21.5 mm, using histological examination. However, the work of Chandler et al. (2011) report the SN to be 29.5 mm in length using immunohistochemical analysis using Cx43 as a marker for the working myocardium (Fig. 1.4). It is noted in this study that the use of a marker in addition to the standard histological analysis is particularly important for identification of nodal tissue at the head and tail, and it could be that it is these areas that are responsible for the differences seen between the studies (Chandler et al., 2011). The size of the SN is variable in the human and does not appear to correlate with heart weight, atrial size or terminal crest thickness (Sanchez-Quintana et al., 2005). The historical guidelines set down by Aschoff and Monkeberg (1910) state that the tissues of the CCS should be insulated from the surrounding myocardium. Histology clearly shows the node to contain a large amount of fibrous tissue that often forms a distinct boundary with atrial myocardium, and it is has been suggested that this fibrous border forms the insulation of the node (Davies et al., 1983). However, not all investigations have shown conclusive evidence that there is an absence of connections between the tissues. There is also evidence for the existence of transitional cells that extend from the SN into the myocardium of the crista terminalis and superior vena cava (James et al., 1966). Radiations of nodal cells have been seen in many human heart studies penetrating up to 2 mm into the atrial myocardium (Davies et al., 1983; Sanchez-Quintana et al., 2005), and it may be that they could act as functional exit pathways from the node (Anderson and Ho, 1998; James, 2001). There is much interest in how the SN activates the surrounding atrial myocardium. Functional recordings of SN activation have been studied in patients. This has shown that far from there being a discrete location for the leading pacemaker site in all hearts, the actual sites involved in initial activation are widespread throughout the intercaval region, with right atrial pacemakers discovered in a region covering 23 Figure 1.4. 3D anatomical model of the human sinus node. From Chandler et al. (2011). 24 7.5 cm by 1.5 cm (Boineau et al., 1988) (Fig. 1.5A). This is a far larger region than even the largest reported anatomical size of the SN in the human. The location of the leading pacemaker site during sinus rhythm is stable with consistent atrial activation maps obtained using a variety of methods including optical mapping (Fedorov et al., 2010), micro electrode recording (Boineau et al., 1988), and ECG P wave analysis (Grossman and Delman, 1969). The site of the leading pacemaker is not static though and has been demonstrated to shift under numerous conditions such as sinus node arrhythmia (Gomes and Winters, 1987; Boineau et al., 1988), vagal stimulation (Gomes and Winters, 1987; Boineau et al., 1988) arrhythmia (Irisawa and Seyama, 1966) and myocardial infarction (Grossman and Delman, 1969). The different regions of the sinus node, from centre to periphery and head to tail, appear to have different pacemaking properties with some parts perhaps functioning as escape pacemakers (Gomes and Winters, 1987). A functional conduction block zone has been shown to exist around portions of the SN, and it has been proposed that there are preferential exit pathways from the SN (Fedorov et al., 2010) (Fig. 1.5C). Human atrial activation was recorded using optical mapping techniques on perfused SN and has shown that during sinus rhythm the site of breakthrough remains stable (Fedorov et al., 2010), suggesting the existence of preferential pathways through the SN, though the breakthrough sites between the samples were variable with superior, inferior and even bifocal breakthroughs shown to exist (Fedorov et al., 2010) (Fig. 1.6). Optical mapping also provides convincing evidence for the existence of a block zone between the SN and the interatrial septum (Fig. 1.6). Anatomically the zone is shown to be made up of the sinus node artery and an area of connective tissue containing few myocytes and fat. After initial activation the atria show preferential conduction along the crista terminalis and into the right atrial appendage (Fedorov et al., 2010) (Fig. 1.6). 1.2.2) Novel paranodal area The study by Chandler et al. (2009) identified an extensive region in the crista terminalis in close proximity to the SN, although not in continuity with it, that consists of loosely packed myocytes. This area was termed the paranodal area (Chandler et al., 2009) (Figs. 1.2 and 1.4). 25 Figure 1.5. Sinus node activation sites and pathways. 26 Figure 1.5. Sinus node activation sites and pathways. A. Sinus rhythm activation maps showing unifocal and multifocal impulse origins. From Boineau et al. (1988). B. Schematic representation of the anatomic distribution of focal atrial tachycardias (black dots). From (Kistler et al., 2006). C. 3D model of human SN, showing multiple layers surrounding SN and functional exit pathways. CT: crista terminalis; IAS: interatrial septum; IVC: inferior vena cava; LAA: left atrial appendage; MV: mitral valve; PV: pulmonary veins; RAA: right atrial appendage; SVC: superior vena cava; TV: tricuspid valve. From Fedorov et al. (2010). 27 Figure 1.6. Optical mapping of sinus node. 28 Figure 1.6. Optical mapping of sinus node. A. Photograph of atrial preparation. SN location shown by pink oval. Sulcus terminalis shown by white dotted line. B. Separated SN and atrial activation maps. Conduction velocities in each direction shown. CT: crista terminalis; IAS; interatrial septum; IVC: inferior vena cava; RA: right atrium; RAA: right atrial appendage; SAN: sinus node; SVC: superior vena cava. From Fedorov et al. (2010). 29 Rather surprisingly, given the distinct morphology of the area, the extent of the paranodal area had not been studied earlier. It has been suggested that the paranodal area is populated by a mixture of atrial and nodal like myocytes. The data for this was obtained through immunofluorescence using two markers, Cx43 and atrial natriuretic peptide (ANP), which are known to be present in the atrial myocardium but absent in the nodal myocytes. The reconstruction in a subsequent study shows the paranodal area to be a more extensive structure than the SN, that appears to extend across a large portion of the crista terminals, and extending at least 5 mm beyond the SN towards the inferior vena cava (Chandler et al., 2011) (Fig. 1.4). It can be seen in the histological sections that the paranodal area is still an extensive structure, and even increasing in size, at the levels of SN head and tail, where the actual node is reducing in size (Fig. 1.3). The inferior extent of the paranodal area cannot be elucidated from this study as it appears to continue past the sampled tissue block. The potential role of the paranodal area is still undetermined; however its key location in close proximity to the SN and in a hotspot for arrhythmogenic foci mean investigation into this region is critical. It may be possible that the paranodal area is responsible for the atrial pacemakers that are found outside of the anatomical SN (Boineau et al., 1988) (Fig. 1.5A). It has also been proposed that the paranodal area may act to facilitate SN function by depolarizing the atrial myocardium surrounding the SN (Chandler et al., 2011). The crista terminalis is a well-known site of origin for focal atrial tachycardias and this form of tachycardia are now commonly termed cristal tachycardias (Kalman et al., 1998). Several studies have reported that sites along the crista terminalis were responsible in around two-thirds of patients seen with right atrial tachycardias (Kalman et al., 1998; Kistler et al., 2006) (Fig. 1.5B). It has been suggested that the crista terminalis may be such a prevalent site for foci due to its close proximity to the SN, and cell-cell coupling within the crista terminalis that favours the formation microreentrant circuits (Kalman et al., 1998). The location of the paranodal area corresponds well with the areas that are suggested as being responsible for these cristal tachycardias, so it seems possible that the paranodal area may provide a substrate for the formation of arrhythmias and may be an important target for their treatment. 30 The study by Chandler et al. (2009) investigated the gene expression of the paranodal area in comparison to the SN and atrial myocardium. It remains unclear whether the paranodal area contains a unique population of cells or whether it is solely an area of mixed population of atrial and nodal myocytes. The fact that the majority of the gene targets appeared to have expression that was intermediate between the two regions could possibly suggest that the paranodal area does not contain a unique cell phenotype. However, mRNA expression for some genes such as Kv4.2 (voltage-gated K+ channel), Kir6.1 (inwardly-rectifying K+ channel), TASK1 (Tandem-pore acid-sensitive K+ channel) and SK2 (small conductance calcium-activated K+ channel) were significantly higher in the paranodal area than with atrium or SN, suggesting that the paranodal may also possess some unique specialised cells. Immunohistochemical analysis of this region using antibodies against Connexin 40 (Cx40), Cx43, ANP and vimentin (an intermediate filament found in fibroblasts) further suggests that the cells present are a mixture of nodal-like and atrial-like myocytes and also that the extracellular matrix in the paranodal area is adipocyte rich rather than fibroblast rich as seen in the SN (Chandler et al., 2011) (Fig. 1.2). 1.2.3) Internodal pathways One of the key remaining controversies concerning the CCS is the potential existence of intra atrial conduction pathways for preferential conduction from the SN to the atrial myocardium and the AVN. Histologically and immunohistochemically there does not currently appear to be strong evidence for the existence of such pathways (Benvenuti et al., 1997; Anderson and Ho, 1998), although their existence is stated in some studies (James, 2001). Mapping of the atria during sinus rhythm does show anisotropic atrial activation (Fedorov et al., 2010) (Fig. 1.6). There are theories that suggest that this preferential conduction, for example along the crista terminalis, may be due to fibre orientation in the atria or differential coupling of atrial myocytes rather than specialised conduction fibres such as the Purkinje fibres found in the ventricles. The propagation of action potentials between the atria has also been shown to occur preferentially through Bachman’s bundle, which is a large muscular bundle with fibres running in parallel along the anterior atrial wall that acts as an interatrial bridge (Anderson and Ho, 1998; Ho and Sanchez-Quintana, 2009). 31 1.2.4) Atrioventricular node In the normal healthy human heart the AVN is the sole conduction pathway connecting the atria and ventricles (Tawara, 2000). The AVN is a slow conducting region, important in creating a delay in action potential propagation, which is critical in ensuring complete filling of the ventricles during diastole before they are stimulated to contract. Without this delay the heart would be unable to pump blood efficiently. As the AVN is the sole pathway for conduction it means that any defects in this area can have serious consequences on cardiac function. There are a number of relatively common pathologies related to AVN dysfunction including heart block and re-entrant tachycardia (Pick and Langendorf, 1974). The location of the AVN in the human heart can be found by identifying the landmarks that form the triangle of Koch. These are the tricuspid valve annulus, Eustachian valve (covering the entrance to the inferior vena cava) extending into the tendon of Todaro and the coronary sinus ostium. The atrial portion of the AVN lies within this triangle before entering the ventricular septum, penetrating the central fibrous body at its junction with the tendon of Todaro (Fig. 1.7A). The AVN is complex structure that can be divided into several distinct portions; the inferior nodal extensions, compact node and His bundle (also known as the penetrating bundle) (Davies et al., 1983). It is the compact node that is considered the true node. Tissue sections through the atrioventricular junction show that the compact node is located towards the right side of the atrioventricular muscular septum, which is formed by the lower tricuspid valve attachment to the septum than the mitral valve (Davies et al., 1983) (Fig. 1.7B). The compact node is separated from the ventricular-septal myocardium by the annulus fibrosus but possesses connections with the atrial myocardium via a zone of loosely packed transitional cells (Fig. 1.7B). Histologically the compact node is seen as a densely packed structure that stains lighter than the surrounding myocardium (Fig. 1.7C). The compact node has been reported to extend for a distance of between 2 and 5 mm (Inoue and Becker, 1998). The penetrating bundle forms at the insertion of the compact node into the central fibrous body. This occurs at the apex of the triangle of Koch. The AVN at this stage is completely insulated from any surrounding myocardium (Fig. 1.7C). In the human left and right inferior nodal extensions have been shown to exist, and extend from the compact node 32 Figure 1.7. Location of the atrioventricular node. 33 Figure 1.7. Location of the atrioventricular node. A. Landmarks of triangle of Koch. From (Anderson and Cook, 2007). B. Histological sections showing location of AVN, and the transition from compact node to bundle branches. From Anderson and Ho (1998). C. Histology of atrioventricular conduction axis from INE (left), CN (middle), PB (left). AM: atrial muscle; CN: compact node; CFB: central fibrous body; INE: inferior nodal extension; PB: penetrating bundle; TA: transitional area; VM: ventricular muscle. From Greener et al. (2011). 34 and project towards the mitral and tricuspid valve annuli (Kurian et al., 2010) (Fig. 1.8). The morphology of these tracts is highly variable between hearts but it is thought that they may function as the input pathways to the compact node portion of the AVN (Inoue and Becker, 1998). The location of these extensions into the triangle of Koch is important as they are often ablation targets in the treatment of reentrant tachycardias (Prystowsky, 1997; Billinton and Knight, 2001). The right extension is consistently seen as the more prominent, with a mean length of 4.4 mm as compared to 1.8 mm for the left extension (Inoue and Becker, 1998). Functional recordings of the atrioventricular junction have revealed the existence of slow and fast pathways to the AVN. The slow pathway runs through the septal isthmus along the base of the triangle of Koch, whilst the fast pathway runs through the atrial septum (Hucker et al., 2008a; Kurian et al., 2010) (Fig. 1.8A). It is thought that the left and right inferior nodal extensions form the anatomical basis for these pathways. The two pathways have distinct electrophysiological properties, with the effective refractory period (the time for which a tissue is unable to be stimulated to initiate a propagated action potential) for the fast pathway being longer (450 msec) than in the slow pathway (340 msec), whilst the functional refractory period (the shortest interval between two conducted impulses resulting from consecutive input impulses) of the fast pathway is shorter than in the slow pathway (Denes et al., 1973). The expression of Cx43 in the pathways has also been shown to be specialized with Cx43 positive slow pathway and negative fast pathway (Hucker et al., 2008b; Kurian et al., 2010) (Fig. 1.8B). A good understanding of the dual pathways of the AVN is important due to their reported role in AV nodal re-entrant tachycardia (AVNRT). AVNRT is caused by conduction abnormalities within the AVN. With normal sinus rhythm atrial action potentials enter the compact node via the fast pathway, and potentials entering via the slow pathway are blocked. Conduction can occur down the slow pathway with a premature atrial action potential blocked in the fast pathway due to its extended refractory period. This leads to a prolonged PR interval on the ECG due to delayed ventricular excitation. A re-entrant circuit forms when slow conduction down the slow pathway retrogradely enters the fast pathway when it is no longer refractory. The impulse can then potentially re-enter the slow pathway, leading to a sustained re-entrant tachycardia. Treatment for AVNRT often involves ablation within the region of the slow pathway, however there often appears to be a mismatch between 35 Figure 1.8. Understanding pathways of the atrioventricular node. A. Structural understanding of atrioventricular junction (AVJ). B. Histological serial sections revealed the presence of rightward and leftward posterior nodal extensions. In addition, immunofluorescence identified differences in the expression of Cx43 between the two extensions and their corresponding counterparts in the AV node. (a) 3D reconstruction of an AVJ. (b–d) Histological sections as denoted in (a) by the solid black lines. (e–g) Corresponding immunostained sections for Cx43. Yellow colour shows Cx43. CFB: central fibrous body; CN: compact node; CS: coronary sinus; IAS: interatrial septum; FP: fast pathway; LE: leftward extension; LNB: lower nodal bundle; RE: rightward extension; SP: slow pathway; TT: tendon of Todaro; VS: ventricular septum. From Kurian et al. (2010). 36 the anatomical and electrophysiological location of the pathways thought to be involved in re-entry (Prystowsky, 1997; Billinton and Knight, 2001), and this has led to different approaches in the clinic with some favouring the anatomical approach of ablation based on the landmarks of the slow pathway and others favouring the functional approach of attempting to identify the slow pathway via endocardial recording (Kalbfleisch et al., 1994; Prystowsky, 1997; Billinton and Knight, 2001). 1.2.5) Ventricular conduction system The left and right bundle branches and the ramifying Purkinje fibre networks form the final portion of the CCS, the His-Purkinje system. They are visible upon opening the ventricular chamber and examining the endocardial surface as pale fibres (Fig. 1.9). This part of the conduction system is insulated from surrounding myocardium by a connective tissue sheath that is only lost at the terminal Purkinje muscle junctions (Ansari et al., 1999; Ghatta et al., 2006). The left and right His-Purkinje networks are asymmetric (Fig. 1.9A). The bundle branches are formed by the branching of the penetrating bundle within the ventricular septum. The bundle branches descend down the ventricular septum subendocardially for approximately 10 to 20 mm before the overlying smooth muscle and collagen are lost (Massing and James, 1976). The right bundle branch is a continuation of the penetrating bundle and exists as an obvious bundle running towards the medial papillary muscle, whilst the left bundle branches off and forms a broad ribbon-like structure that runs down the left ventricular septum (Davies et al., 1983; Tawara, 2000). The human left bundle branch is said to be a trifascicular structure with projections towards the papillary muscles and towards the apex of the heart (Massing and James, 1976; Tawara, 2000; Sawaya et al., 2012). The Purkinje fibres are highly specialised fibres for the rapid conduction of action potentials and are direct continuations of the bundle branches. The Purkinje fibre networks in both ventricles divide and coalesce extensively forming complex 3D networks of fibres, often as free-running fibres or false tendons (Tawara, 2000). The Purkinje fibres networks appear to project predominantly towards the papillary muscles. The free running Purkinje fibres reattach to the ventricular myocardium and project into the endocardium, where they lose their connective tissue sheath and form connections allowing action potential propagation to the ventricles. 37 Figure 1.9. Anatomy of the human His-Purkinje system. A. Left and right HisPurkinje networks as demonstrated by Tawara in 1906. From Tawara (2000). B. Photograph of left ventricle of human heart showing portion of bundle branch and ramifying Purkinje network. 38 The Purkinje fibres are required in order to ensure that contraction of the ventricles occurs from the apex to the base of the heart. This is in order to achieve the most efficient pumping of blood out of the heart. It is possible for re-entrant circuits to form both between the left and right bundle branches and also between the fascicles of the His-Purkinje network, in a similar manner to re-entry in the AVN. There is also some evidence of the Purkinje fibres acting as the source of ectopic foci that can trigger and sustain ventricular arrhythmia such as ventricular tachycardia (Nogami, 2011a; Nogami, 2011b). The potential pacemaking capacity of the Purkinje fibres becomes more prevalent during myocardial infarction, thought to be due to removal of overdrive suppression from the SN. It has been suggested that this is due to the Purkinje fibres being resistant to hypoxia due to their high glycogen content and cavital blood supply, and can lead to polymorphic ventricular tachycardia after myocardial infarction (Nogami, 2011b; Carvalho-de-Souza et al., 2013). Catheter radiofrequency ablation treatment of Purkinje related arrhythmias, such as ventricular fibrillation, is guided by 3D activation mapping of the ventricular septum, but it is critical to ensure that bundle branch block is not induced (Nogami, 2011a; Nogami, 2011b), and therefore a good understanding of the complex anatomy of the His-Purkinje network is important. Bundle branch block can occur in both the left and right networks. Right sided bundle branch block is often thought to be benign but left bundle branch block often leads to serious cardiac dysfunction with reduced cardiac ejection fraction (Brenner et al., 2000; Breithardt and Breithardt, 2012). 1.3) Analysis of existing cardiac models A number of different methodologies have been implemented in the construction of 3D models of the human cardiac anatomy and also the human CCS. Historically, reconstructions were based solely on serial histological sections that were then manually segmented and overlaid onto images of the cardiac anatomy in order to guide the reconstruction (eg. Truex and Smythe, 1967; Truex et al., 1967). More recently groups have been using high resolution magnetic resonance imaging (MRI) and computed tomography (CT) in order to obtain detailed 3D data for anatomy (Chandler et al., 2011; Deng et al., 2012) (Figs. 1.4, 1.10). 39 Figure 1.10. Models of human cardiac conduction system and activation. 40 Figure 1.10. Models of human cardiac conduction system and activation. A. Human heart conduction system. (a) posterior view of the atria; (b) conduction bundles in the atria; (c) transparent display of the conduction bundles and atrial muscles; (d) posterior view of the ventricle; (e) conduction bundles in the ventricle; (f) transparent display of conduction bundles and ventricular muscles. AVR: atrioventricular ring; IVC: inferior vena cava; LAM: left atrial myocardium; LBB: left bundle branch; LHIS: left His bundle; LP: left Purkinje fibre system; PV: pulmonary veins; RAM: right atrial myocardium; RBB: right bundle branch; RHIS: right His bundle; RP: right Purkinje fibre system; SAN: sinus node. From Deng et al. (2012). B. Example of a 3D volumetric adjustment of conductivity in the heart with scar; with model initialization (d0-map and pacing location) and final results (d1map) and associated mean error in activation times after adjustment. From (Pop et al., 2012). 41 However the resolution obtained from both of the systems is insufficient to allow the accurate determination of the location of the CCS in the hearts. Ambrosi et al. (2009) used optical coherence tomography (OCT) to perform virtual histology on the human heart (Fig. 1.11) and whilst it provided images of sufficient quality to identify structures based on their anatomy it still lacks the ability to positively label for the conduction system (Ambrosi et al., 2009). Currently this can only be accurately achieved through the use of histology and immunohistochemistry. Chandler et al. (2011) used an incorporated method of overlaying histology and immunohistochemistry images onto diffusion tensor magnetic resonance imaging (DT-MRI) data obtained for the SN prior to sectioning, allowing the SN to be accurately identified and outlined and also accurately positioned within the cardiac anatomy. This method also allowed the determination of myocyte orientation that was also incorporated into the model (Chandler et al., 2011). It is common for many cardiac reconstructions, particularly for the human due to its large size, to focus on only one component which may lead to errors if two models created using different methods are subsequently consolidated into a single model. Deng et al. (2012) created a detailed 3D reconstruction of the whole human cardiac anatomy including major landmarks and fibre orientation. However, the CCS was created from data published from other hearts rather than specifically from the heart that was being reconstructed (Deng et al., 2012). Again this may lead to errors in modelling studies. Stephenson et al. (2012) used contrast enhanced micro-CT to reconstruct the CCS in the rabbit heart, based on the differing attenuation properties of the CCS tissues compared to the working myocardium. This produced a very accurate and detailed representation of the whole CCS but again lacks the ability to use a marker for the CCS, meaning that there may be additional components that are missed in this form of modelling. Computer modellers require accurate cardiac anatomy mathematical arrays onto which they can incorporate action potential characteristics, for them to accurately simulate cardiac activity (Seemann et al., 2006; Aslanidi et al., 2011). It is important that the anatomical models are as extensive and accurate as possible as this increases the usefulness of the model to examine processes across the whole heart. There has been limited functional work carried out on isolated human hearts and tissue preparations. This makes validation of computer models difficult and it is 42 important for the advancement of modelling that accurate clinical data is obtained, however there have been some steps taken towards integrating optical mapping data with 3D reconstruction (Fedorov et al., 2010; Pop et al., 2012) (Fig. 1.10B). 43 Figure 1.11. Comparison between histology and OCT. Left panels. Histology (top) and corresponding OCT (bottom) of sinus node. (*) identifies sinus node. CT: Crista Terminalis; SN: Sinus node. Right panels. Histology (top) and corresponding OCT (bottom) of infarcted right ventricle. Dotted line represents infarct border. From Ambrosi et al. (2009) 44 CHAPTER 2) GENERAL METHODS 2.1) Human sample details and ethical approval A human sinus node preparation and sinus node tissue sections from unused donor hearts from patients with no history of heart disease were obtained from Prince Charles Hospital, Queensland, Australia, from Professor Peter Molenaar. Ethical approval for use of this human tissue was provided by the Prince Charles Hospital (EC2565). A whole intact human heart was obtained from University of Minnesota, USA, from Professor Paul Iaizzo. Ethical approval was provided by the University of Minnesota (9901M00097). All work was carried out in accordance with the Human Tissue Act (2004). Patient details for all samples used are provided in Table 2.1. 2.2) General principles 2.2.1) Immunohistochemistry 2.2.1.1) Principles Immunohistochemical staining techniques on tissues have been in us since the late nineteenth century. The principal revolves around the use of antibodies to specific antigens in order to visualise their localisation within a tissue. In order to visualize the initial antigen antibody reaction a secondary antibody conjugated to a reporter such as a fluorophore or biotin is used. Classes of antibodies – Antibodies used for immunohistochemistry are typically IgG or IgM. Antibodies are glycoproteins consisting of two heavy and two light polypeptide chains. IgG antibodies are a monomer arranged in a “Y” shape with antigen binding sites on the 2 arms (Fragment, antigen binding region, Fab) and functional sites on the tail portion (Fragment, crystallisable region, Fc). IgM antibodies consist of 5 or 6 linked “Y” shaped subunits, each of which are comprised 45 Tissue Patient 1 Aetiology Sex Age - - - - 2 Donor F 30 3 Donor M 31 4 Donor Cystic fibrosis - Non failing heart - heart lung liver transplant Sub arachnoid haemorrhage with cardiorespiratory arrest Cerebral haemorrhage F 40 5 Donor - - - 6 Donor Sub arachnoid haemorrhage - brain death F 40 7 Donor - - - Ethics Table 2.1. Heart details 46 KCa Immuno KCa qPCR 3D model 9901M00097 Whole heart EC2565 (Australia) EC2565 (Australia) EC2565 (Australia) EC2565 (Australia) EC2565 (Australia) EC2565 (Australia) Right atrium Left ventricle Sinus node and adjacent atrial muscle Sinus node and adjacent atrial muscle Sinus node and adjacent atrial muscle Sinus node and adjacent atrial muscle of 2 heavy and 2 light polypeptide chains, and possessing Fab sites, joined by a J chain (Fig 2.1). Antibodies produced for immunohistochemistry are classified as either monoclonal or polyclonal depending on the methods used for their production. Polyclonal antibodies are created by injecting a host with a specific antigen. This leads to the creation of specific IgG antibodies in the host as part of the normal immune response. Serum from the host can then be extracted and the antibodies it contains purified. The purified antibodies consist of a mixture of different antibodies produced from numerous different immune cell types, but all specific for the same antigen, therefore the antibodies are termed polyclonal. Large animals such as the goat are typically used as hosts for the production of polyclonal antibodies in order to obtain large volumes of serum from which to obtain the antibodies, but rabbit polyclonal antibodies have been reported to offer greater antigen recognition (Buchwalow and Böcker, 2010). Monoclonal antibodies are produced by a single cell type. Typically it involves the injection of a host with a specific antigen, followed by the collection of immune cells from the spleen. These cells are then used in the creation of hybridomas, which are formed by joining tumour cells to the antibody-producing mammalian cells. The hybridomas can then be grown in culture or injected into a living host in order to continually produce specific antibodies. For monoclonal antibodies mice are normally used. Secondary antibodies are used to visualize the location of the bound primary antibodies. Secondary antibodies are produced in a similar fashion as for primary antibodies; however, once the antibody has been purified it is covalently conjugated to a reporter such as a fluorescent dye. The secondary antibodies are species specific and bind directly to the primary antibody. This method produces an indirect visualization of the target antigen. 2.2.1.2) Protocol steps Tissue fixation – Tissue fixation is a critical step in immunohistochemistry and can greatly influence the quality of results obtained. Adequate fixation to preserve the tissue needs to be achieved whilst preserving its antigenicity and avoiding damaging 47 Figure 2.1. Basic antibody structure. Top. IgG. Bottom. IgM. Antibodies consist of heavy and light polypeptide chains joined together by disulphide bonds. Each ‘Y’ shaped fork possesses two identical antigen binding sites. IgG antibodies are a monomer arranged in a “Y” shape with antigen binding sites on the 2 arms. Functional sites are found on the tail portion. 48 the morphology of the tissue sections. There are two general fixation methodologies that are widely used, using cross-linkers or denaturation using organic solvents. Cross-link fixation involves the chemical addition of fixing agents to proteins and cellular components and the formation of inter-molecular and intra-molecular crosslinks, for example formaldehyde reacts extensively with amino groups to form methylene bridges (Rolls, 2012). Denaturation fixation involves the replacement of water in tissues with an organic solvent such as ethanol or methanol. This leads to an alteration of the structure of proteins due to a change in hydrophobic bonds making them water insoluble. Cross-link fixation is generally preferred as it maintains the integrity of the tissue morphology better than denaturation with less cell shrinkage observed and less antigen blocking (Boenisch, 2005). However cross-link fixation has been linked to antigen masking brought about due to the conformational change of the protein structure altering the antigen binding site of antibodies (Boenisch, 2005). Permeabilization – Detergents can be used in order to increase the permeability of the cell membrane to antibodies therefore increasing antibody penetration. Triton X100 is a frequently used detergent but as it damages the membrane is its use is not advised for antigens that are expected in the cell membrane. Blocking – For clear visualization of immunofluorescence staining background signal needs to be kept to a minimum. The background signal can originate from numerous sources. For example it is possible for Fc receptors in the cell membranes to bind the Fc domain on antibodies leading to non-specific labelling (Buchwalow and Böcker, 2010). These receptors can be blocked by using solutions containing bovine serum albumin (BSA) or normal serum from the host of secondary antibody. Also all tissues have a degree of autofluorescence due to substances such as lipofuscins, porphyrins, elastin and collagen (Buchwalow and Böcker, 2010). Heart tissue has been shown to contain a large number of autofluorescent lipofuscin molecules and the quantity increases with age and in conditions in disease such as ischemia (Billinton and Knight, 2001; Majno and Joris, 2004). These molecules can mask specific antibody staining and requires that immunofluorescence images are carefully analysed to ensure observed signal is a true and accurate representation of the location of the antigen being investigated. 49 Washing – Phosphate buffered saline (PBS) with a pH of 7.4 is commonly used to wash tissue sections between the different stages of the immunohistochemistry protocol. Primary antibody incubation – Primary antibodies are diluted to a working concentration in a solution of 1% BSA in PBS (pH7.4). The optimal concentration for primary antibodies can be extremely variable and needs to be optimised at the outset of the experiment. An initial experiment can be performed using serial dilutions of primary antibody ranging from 1:50 to 1:800 to optimise signal intensity whilst keeping background staining to a minimum. For human tissue, sections are incubated overnight at room temperature. Primary antibodies used are listed in Table 2.2. Secondary antibody incubation – Secondary antibodies are prepared to working concentrations in a solution of 1% BSA in PBS (pH7.4). Sections are incubated at room temperature for 2 hours in the dark in order to prevent photo-bleaching of the fluorophore. Secondary antibodies are selected that are specific for the species and form of the primary antibody. The secondary antibodies are conjugated to a fluorophore that when excited at a specific wavelength emits a specific wavelength light (Table 2.3). The optimal concentration of secondary antibody can be affected by the fluorophore selected and the primary antibody antigen. Secondary antibodies used are listed in Table 2.4. Mounting – Anti-fade and anti-photobleaching aqueous mounting medium needs to be used for immunofluorescence slides in order to preserve the fluorescence signal so that the slides can be viewed over a longer period of time and also to protect the fluorophore when it is exposed to an excitation signal. Vectashield H-1000 mounting medium (Vector Labs) is a commercially available mounting medium optimized for immunofluorescence. After a coverslip has been placed on the slide it needs to be sealed using nail varnish to retain the aqueous mounting medium. Negative controls – It is important to assess the level of autofluorescence in a tissue caused by the staining protocol in comparison to the positive experiments. Negative control experiments are performed where the primary antibody is omitted and instead the sections are incubated in 1%BSA. 50 Protein Host HCN4 Rabbit Polyclonal RYR2 Mouse Cx43 Caveolin Vimentin Smooth muscle α actin KCa1.1 (intracellular, 1097-1196) KCa1.1 (intracellular, 1184-1200) KCa1.1 (extracellular, 199-213) Type Concentration Manufacturer IgG 1:50 Alomone Labs Monoclonal IgG 1:100 Thermo Scientific Mouse Monoclonal IgG 1:50 Millipore Mouse Guinea Pig Monoclonal IgG 1:50 BD Biosciences Polyclonal IgG 1:100 Progen Mouse Monoclonal IgG 1:100 Sigma Aldrich Rabbit Polyclonal IgG Range 1:50 – 1:800 Alomone Labs Rabbit Polyclonal IgG Range 1:50 – 1:800 Alomone Labs Rabbit Polyclonal IgG Range 1:50 – 1:800 Alomone Labs Table 2.2. List of primary antibodies used 51 Fluorophore Cy3 (Cyanine 3) FITC (Fluoresceinisothiocyanate) Excitation wavelength (nm) 552 Emission wavelength (nm) 565 Colour 490 520 Green Red Table 2.3. Excitation and emission wavelengths of secondary antibody dyes 52 Table 2.4. List of secondary antibodies used Host Type Conjugate Concentration Manufacturer Goat Rabbit IgG FITC 1:100 Millipore Donkey Rabbit IgG FITC 1:100 Millipore Goat Mouse IgG Cy3 1:400 Millipore Donkey Mouse Guinea pig IgG Cy3 1:400 Millipore IgG Cy3 1:400 Millipore Donkey 53 2.2.2) Laser Scanning Confocal Microscopy Principles Immunofluorescence slides can be visualized using confocal microscopy. Confocal microscopy involves the principle of targeting tissue with a specific wavelength of light created by a laser and then eliminating out of focus light emitted from the tissue by using a pinhole in front of the detector. This enables high resolution and contrast images to be obtained. A schematic diagram of a confocal microscope is shown in Figure 2.2. Laser – Helium-neon (HeNe) and argon-ion lasers are used to generate monochromatic light. Individual lasers are required to generate light with different wavelengths and so the lasers available on a confocal microscope setup needs to be assessed when choosing fluorophores for immunofluorescence experiments. Beam Splitter – The light generated by the lasers passes through a dichromatic beam splitter which separates light of long and short wavelengths. The short wavelength light is reflected towards the objective lens whilst light of longer wavelengths passes through the splitter. Scanning unit/head - A scanning unit moves a focused laser beam over the tissue. Objective lens – A confocal microscope setup uses epi-illumination, where the objective lens acts to focus the excitation light and the emission light. Pinhole – The pinhole lies in front of the photomultiplier unit. By adjusting the aperture diameter it is possible to prevent out of focus light from reaching the photomultiplier. The diameter of the aperture determines the thickness of tissue from which the emitted light is collected from. The smaller the diameter, the thinner the optical slice thickness and the greater depth discrimination of fluorophore location within a tissue section. However the trade off with a small aperture diameter is a reduction in signal which can limit the detection of the fluorophore. Photomultiplier (PMT) – The photomultiplier collects the emitted light that passes through the pinhole converting the analogue signal into a digital signal. The PMT is a photocathode tube and creates pixel values based on the number of photons absorbed by the photocathode allowing the signal strength to be collected. 54 Figure 2.2. Major components and principle of a confocal microscope. The laser creates a beam that is focused onto a beam splitter which only allows light of specific wavelength to pass through and reflects the remaining light. The light reflected by the beam splitter is then focussed onto the tissue by the objective lens. Light emitted from the excited fluorophores passes back through the objective lens. This light can now pass through the beam splitter as the emission wavelength is longer than the excitation wavelength. A pinhole sits in front the signal detection system and operates to prevent out of focus light reaching the detector. photomultiplier detects the signal and converts it into a digital signal. 55 A 2.2.3) Real-Time Quantitative PCR The polymerase chain reaction (PCR) allows the amplification of DNA. By incorporating a fluorescent reporter into the reaction it is possible to assess the quantity of DNA present in real time leading to the technique of real-time quantitative PCR. 2.2.3.1) Principles A PCR reaction must contain specific components, the final concentration of which can greatly affect the quality and efficiency of the PCR reaction. A thermostable DNA polymerase enzyme, such as Taq polymerase, is required for the DNA synthesis, whereby nucleotides are added to the 3’ end of the growing strands. The nucleotides required for this amplification come from the addition of free deoxynucleotide triphosphate molecules (dNTPs) into the reaction mix. MgCl2 is required in the reaction in order to provide Mg2+ which acts as a co-factor for the DNA polymerase. Buffers are also required in order to maintain optimal pH conditions. In order for amplification to occur short priming sequences are required, which can either be specifically designed, in order to amplify a specific target sequence or random primers can be used, in order to amplify the whole DNA strand. PCR involves the heating and cooling of samples to specific temperatures. The heating of the reaction mix to 95oC leads to the separation of double stranded DNA by breaking hydrogen bonds between complementary base pairs on DNA strands. When the sample is then cooled to 55oC the free primers are able to anneal to the single stranded DNA. Then when the sample is heated up to 72oC the DNA polymerase extends the DNA strands, leading theoretically to a doubling of the DNA present in the reaction. This heat cycling process can be repeated in order to further amplify the DNA. qPCR requires complementary DNA (cDNA) as an initial input product. Therefore after the RNA isolation it is required to form cDNA from the RNA template, so called first strand synthesis. This is achieved through reverse transcription using a reverse transcriptase enzyme (RNA-dependent DNA polymerase) and typically random hexamer or oligo-dT primers, in order to form a DNA-mRNA double stranded hybrid molecule. The RNA is subsequently degraded using RNase H, leaving a DNA strand. 56 It is possible to quantify the amount of RNA in a solution by using a spectrophotometer. Both RNA and DNA absorb light with a wavelength of 260 nm, allowing the amount present in solution to be calculated. However as both RNA and DNA affect the absorbance it is important to have a pure RNA sample in order to obtain an accurate measurement. By also measuring the absorbance at 280 nm and 230 nm and comparing it to the 260 nm reading it is possible to gain an indication of the quality of the sample and potential presence of contaminants. Primers Primer annealing to single strands occurs at a temperature of around 55oC, but at this temperature the complementary DNA strands present in the reaction can also start to anneal to each other, therefore a high concentration of primers is used in order to ensure primer binding each cycle. Primer specificity is critical in order to ensure that only the sequence of interest is amplified. To avoid the problem of genomic DNA contamination primers are typically designed against part of the sequence that spans an intron exon boundary, and therefore will be unable to bind to genomic DNA and so no amplification will occur, and so the product obtained will have originated from the RNA template. Quantitative PCR In order to assess the amount of DNA within a reaction a suitable reporter has to be used. For real-time qPCR a fluorescent dye is typically used. One such dye is SYBR green, which is an intercalating fluorescent dye that fluoresces when bound to double-stranded DNA (Fig 2.3). Therefore the more double stranded DNA present in a reaction the larger the fluorescent signal that will be detected. However as SYBR green binds to any double stranded DNA care has to be taken that only the specific target sequence is being amplified in the reaction. A standard qPCR reaction involves 40 thermocycles with a fluorescent measurement taken during each cycle. When the reaction is 100% efficient there is expected to be a doubling of the quantity of DNA per cycle. When the fluorescence measured in a reaction is plotted over the 40 cycles there are distinct phases that can be observed (Fig 2.4A). Initially there is a baseline phase, where the fluorescence detected tends to be masked by noise within the system. 57 Figure 2.3. Principle of SYBR green PCR quantitation. SYBR green fluoresces when bound to double stranded DNA. As DNA polymerization occurs more SYBR green can be bound thereby increasing the fluorescence signal, which can then be quantified. Adapted from Biosyn.com (2014). 58 Plateau phase Exponential phase Ct threshold Background Figure 2.4. qPCR data analysis. 59 Figure 2.4. qPCR data analysis. A. Amplification curve. Distinct phases are observed in amplification curves. Background phase is due to low-level signal. The threshold limit is set in the exponential amplification phase. The cycle number when a reaction reaches this threshold determines the Ct value. The exponential phase is when the reaction is running at its maximum efficiency. The plateau is when the reaction is no longer amplifying efficiently due to reagents being diminished. Melt curve. B. Different PCR products can be distinguished by their melting properties, which alters with product size and nucleotide composition. A single melt curve peak suggests the presence of a single PCR product. 60 This is followed by the start of the exponential phase where the fluorescence increases exponentially. As the exponential phase progresses, the fluorescence plot enters a linear phase, where amplification is occurring at the maximal rate. Finally there is a plateau phase, which is where the reaction becomes limited due to a shortage of components or the presence of large amounts of DNA can interfere with the reaction. For analysis it is the linear phase that is important. A threshold fluorescence value is determined and then the cycle number at which the fluorescence within each sample reaches the threshold is calculated. This number is called the Ct value. The efficiency (Eff) of PCR reactions can be calculated by determining the change in fluorescence (F) observed around the Ct value. Eff = √FCt+1/FCt-1 In order to analyse qPCR results the fluorescence signal obtained for each sample from an endogenous control gene (a gene whose expression is known to be the same in all samples and is unaltered by all of the experimental conditions being tested) is required. By normalising results to the endogenous control it is possible to account for any slight variations brought about due to different quantities of cDNA being loaded into each reaction, meaning any differences observed should be solely caused by the experimental conditions. The normalised data provides the ΔCt value. ΔCt = Eff Ct(control)/Eff Ct(target) 2.2.4) Histology 2.2.4.1) Principles - What is histology Histology is the use of stains in order to visualize structures in thin sections of tissues. The staining of tissue sections allows their morphology to be assessed and the presence and location of particular molecules to be determined. Stains can be used in isolation or in combination in order to enable multiple components to be visualized such as nuclei, collagenous connective tissue, and myocytes. Most typical dyes work by the ionic binding, for example between negatively charged groups on dyes to positively charged amino groups on the tissue. The pH of 61 dye solutions is critical to their function and commonly dyes have a pH in the region of 1.5 to 3 (Bancroft and Gamble, 2008). Histological stains can be classified as either being progressive or regressive in nature. Regressive stains require the final level of staining to be achieved by differentiation, the washing out of excess stain, whereas progressive staining is achieved via application of a weaker stain until the required level of staining is achieved. Bouins fluid is a common fixative used in histology as it acts as a mordant thereby enhancing the intensity of subsequent stains. 2.2.4.2) Common Stains Two of the most common staining combinations used are Masson’s trichrome, which uses celestine blue, hematoxylin, acid fuchsin and methyl blue stains, and hematoxylin and eosin (H&E). These staining protocols use combinations of various stains in order to identify particular cell types. Celestine blue is an acid resistant nuclear stain and is used to preserve nuclear staining throughout the rest of the protocol. The nuclei are stained blue/black. Mayer’s hematoxylin is a progressive general purpose nuclear stain. Chromatin in the nuclei is stained blue/black. Acid fuchsin is an acidic dye that is used to stain myofilaments red. Phosphomolybdic acid acts to displace stain that is already bound to tissue. Collagen is destained more quickly than the myocardium therefore additional stains can be used subsequently to stain the collagen. Methyl blue stains collagen fibres royal blue. Eosin is a general purpose cytoplasmic stain. It stains both myocytes and collagen pink. 62 2.2.5) Micro-CT 2.2.5.1) Principles Computed tomography is a form of X-Ray imaging. X-Ray imaging works on the principle that tissues of differing densities will differently attenuate x-rays, the more dense an object the darker it appears as more X-Rays are attenuated, thereby allowing the visual separation of structures based on their attenuation properties. Using a single X-Ray projection it is only possible to visualize objects in 2D, and information is lost as all objects that have been irradiated contribute to the final image. However, if a large number of X-Ray projections are collected over at least 180o it is possible to reconstruct the data using mathematical reconstruction algorithms in order to generate a 3D geometry. It is then possible to visualize all of the individual features that are present in each of the individual projections. CT scanners are comprised of an X-Ray source, a sample stage and a detector. Typically projections are collected over 360o with either the sample being rotated with a fixed X-Ray source and detector or by rotating the X-Ray source and detector (Fig. 2.5). 63 Detector Figure 2.5. Principle of computed tomography. X-Rays are used to image a sample. A circle scan trajectory is used in order to collect projections over 360o. These scans can then be reformed in order to provide the tomographic images. 5 projections are shown in this image to show how areas within the tissue will be detected from different projections. Adapted from (Semmler and Schwaiger, 2008) 64 CHAPTER 3) 3D ANATOMICAL RECONSTRUCTION OF THE COMPLETE HUMAN SINUS NODE AND PARANODAL AREA 3.1) Introduction 3D reconstruction of the SN in the human heart have been created based on data obtained from several techniques including immunohistochemistry (Chandler et al., 2011). MRI, CT, histology and These techniques have clearly demonstrated that in the human the SN is a crescent shaped structure that is located towards the epicardial surface of the right atrium in the intercaval region. The paranodal is an area of loosely packed myocytes identified in close proximity to the SN. The paranodal area has been shown to contain a mixture of nodal like and atrial like myocytes based on immunohistochemical experiments. Previous reconstruction of the paranodal area in the human heart has shown that it extends towards the superior vena cava, towards a level similar to the SN. The inferior extent of the paranodal area has not previously been investigated. The morphology of the paranodal area in the model created by Chandler et al. (2011) suggests that it is likely to extend further inferiorly within the crista terminalis. 3.1.1) Aim The aim of this study was to construct a 3D model of the complete intercaval region of the human right atrium in order to reveal the morphology of the SN and paranodal area, their relationship to each other, and reveal the full extent of both of these structures within the right atrium. 3.2) Materials and methods 3.2.1) Sample preparation The SN preparation was fixed in 10% neutral buffered formalin (pH 7.4; Sigma) for two weeks prior to CT scanning. After scanning the sample was snap frozen in isopentane cooled in liquid N2 (Fig 3.1). The sample was then stored at -80oC. 65 SVC SVC CT RA IVC IVC Figure 3.1. Sinus node sample. Sample used for reconstruction of sinus node and paranodal area. Sectioning plane is shown by dashed line. CT: crista terminalis; IVC: inferior vena cava; RA: right atrium; SVC: superior vena cava. 66 3.2.2) X-Ray Computed Tomography Computed Tomography (CT) scanning was carried out using a Nikon Metris Custom Bay at the Manchester X-Ray Imaging Facility, University of Manchester. The sample was cleared of fixative by washing in distilled water and then immobilized in a holder to ensure there would be no movement during the imaging process. The scan was acquired with X-ray conditions of 85kV and 180µA. 2100 projections with an exposure time of 2s were collected over 360o. The human SN sample was scanned at a resolution of 31.1µm. 3.2.3) Tissue sectioning After CT scanning the SN preparation was sectioned at a thickness of 25 µm onto SuperFrost Plus slides (VWR) using a Leica CM3050 S cryostat (Leica Microsystems). Five serial sections were collected and then 500 µm was discarded (forming each level) until the whole preparation was sectioned. 3.2.4) Histology One slide from each level (625µm) was selected for Masson’s trichrome histology. Masson’s trichrome histology stains were prepared in accordance with protocols in Bancroft and Gamble (2002). Slides were fixed in Bouin’s fluid (Sigma Aldrich) overnight and then cleared using three 10 minute washes in 70% ethanol. Slides were stained with celestine blue (Sigma Aldrich) for 5 minutes, rinsed in distilled water, stained with Mayer’s hematoxylin (Sigma Aldrich) for 10 minutes, washed in tap water for 15 minutes, stained with acid fuchsin (Sigma Aldrich) for 4 minutes, then rinsed in distilled water until no stain leached from sections. The slides were then stained with phosphomolybdic acid for 5 minutes (Sigma Aldrich) followed by methyl blue (Sigma Aldrich) for 5 minutes, and then rinsed in distilled water. Slides were then treated with 1% acetic acid for 2 minutes, followed by dehydration with ethanol, using 70% for 1 minute, 90% for 1 minute and 100% ethanol twice for 2 minutes. Finally the slides were washed twice in Histo-Clear (National Diagnostics) for 5 minutes to clear the dehydrant, and mounted with coverslips using DPX mountant (Sigma Aldrich). 67 Sections were imaged using Zeiss SteREO Discovery.V8 (Carl Zeiss Microscopy) and Zeiss Imager.Z1 microscopes (Carl Zeiss Microscopy) using Axiovision software (Carl Zeiss Microscopy). With this technique, connective tissue was stained royal blue, cardiac myocytes were stained pink and nuclei were stained dark blue (Fig. 3.2). 3.2.5) Immunohistochemistry Adjacent slides to those selected for histology were used for immunohistochemistry. Tissue sections were fixed in 10% neutral buffered formalin (Sigma Aldrich) for 30 min and then washed three times (10 min each) in 0.01M phosphate buffered saline (PBS) containing NaCl 0.138 M, KCl 0.027 M, pH 7.4 (Sigma Aldrich). The sections were permeabilized by treatment with 0.1% Triton-X100 (Sigma Aldrich) in PBS for 30 min followed by three PBS washes (10 min each). Sections were blocked using 1% bovine serum albumin (BSA) (Sigma Aldrich) in PBS for 60 min. The sections were incubated in rabbit polyclonal anti-HCN4 (Alomone Labs) and mouse monoclonal anti-Cx43 (Millipore), prepared at a concentration of 1:50 in 1% BSA overnight at room temperature. Sections were washed three times in PBS (10 min each) and then incubated in Cy3 conjugated donkey anti-mouse IgG (Millipore) and FITC conjugated donkey anti-rabbit IgG (Millipore) secondary antibodies prepared at concentrations of 1:400 and 1:100 in 1% BSA respectively for 2 hours. Sections were washed three times in PBS (10 min each) and mounted in Vectashield mounting medium (Vector Laboratories). Sections were imaged using Zeiss LSM5 laser scanning confocal microscope (Carl Zeiss Microscopy) using Pascal software (Carl Zeiss Microscopy). 3.2.6) 3D anatomical reconstruction An overview of the 3D reconstruction workflow is shown in Figure 3.3. CT data imported as an image stack into Avizo v5.2 (Visualization Sciences Group, France) and an Isosurface created. ObliqueSlice module used to determine the actual plane in which the tissue block was sectioned and obtain the CT images that corresponded to the histological levels. Histology images from each level were aligned with CT images using Photo-Paint (Corel) in order to account for tissue deformation that occurs during the tissue sectioning process. The locations of SN and paranodal area were identified based on histological and immunohistochemical criteria. immunohistochemistry the SN is an area that 68 Using Epicardium Endocardium Figure 3.2. Histology of sinus node sample. PA: paranodal area; SN: sinus node; SVC: superior vena cava. 69 Figure 3.3. Sinus node reconstruction workflow. A. Sample CT scanned. B. Sample sectioned and histology performed to identify sinus node and paranodal area. C. Immunohistochemistry using HCN4 and Cx43 antibodies used to confirm sinus node identification. D. CT, histology and immunohistochemistry images merged and identified structures outlined. E. Structures to be reconstructed filled. F. Final image used for 3D reconstruction using Avizo. 70 is HCN4 positive and Cx43 negative, and histologically it is seen as an area of compact myocytes located within an area of connective tissue. The SN, paranodal area and atrial tissue were outlined using Photo-Paint (Corel). 47 outlined images were imported into Avizo using Stacked-Slices module allowing correct intervals between levels to be entered (Fig. A1). The ChannelWorks module was used to create single channel black and white images suitable for segmentation. Images were manually aligned with each other prior to segmentation using AlignSlices module. Image segmentation of each region of interest performed on each image to assign labels for each tissue type. Segmented data was visualized using SurfaceGen module. Unconstrained smoothing was applied to the surface. 3.3) Results A 3D anatomical reconstruction on the human SN and paranodal area was created (Fig. 3.4, Movie 3.1). The dimensions of the reconstructed structures are given in Table 3.1. The morphology of the reconstructed SN is similar to that seen in previous anatomical studies of the human SN (Matsuyama et al., 2004; SanchezQuintana et al., 2005; Chandler et al., 2011). Overall the reconstructed SN was 1.6 cm in length. The SN has a broad body region and narrow head and tail regions. The body of the SN had a maximum width of 5 mm and height of 2.3 mm. The head of the SN was 2.3 mm high and 1.8 mm wide. The SN tail was 1 mm high and 2.4 mm wide (Table 3.2). The SN was located predominantly towards the epicardial surface of the right atrium and possessed a crescent shaped morphology. It appeared to follow the line of the sulcus terminalis. The SN was closest to the epicardial surface during its body portion. The head and tail of the SN are located deeper in the atrial myocardium. From the histology there appears to be several distinct regions where the SN projects into the atrial myocardium. These appear as spike like extensions (Fig 3.5). The paranodal area can be seen to be an extensive structure approximately 2 cm in length, situated within the crista terminalis that extends beyond the inferior extent of the SN towards the inferior vena cava (Fig. 3.4). 71 Figure 3.4. Sinus node anatomical reconstruction. A. Epicardial view. B. Endocardial view. C. Side view. SVC: superior vena cava 72 Figure 3.5. Sinus node extensions. Extensions from the sinus node are seen projecting into the atrial myocardium. 73 Material Atrial tissue Sinus node Paranodal area Total Triangles 2167632 16128 48480 2232240 Volume 1567.8 11.46 32.82 1612.08 Table 3.1. Sinus node reconstruction statistics. The number of triangles used to form the surface of each structure and the final volume of each tissue is given. 74 This study Chandler et al., 2011 Matsuyama et al., 2004 Length (cm) 4.6±1 Crista terminalis 3.2 4.0 Sinus node 1.6 2.95 2.1±0.7 Paranodal area 2 3.24 n/a SanchezQuintana et al., 2005 2-3 1.35 (0.82.15) n/a Table 3.2. Comparison of sinus node sizes. A comparison of the sinus node and paranodal area sizes observed in this study as compared to three previous studies on the human heart. 75 It possesses an ellipsoid morphology, being broadest at its mid-point. The superior extent of the paranodal area could be traced to a level corresponding to the mid portion of the SN. A distinct paranodal area could not be observed in the region of the head of the SN. From the reconstruction it appears the SN and paranodal area overlap only for around 7.5 mm. Whilst there is a close proximity between portions of the SN and paranodal area there was no evidence of any direct connections between the two regions in this study and as has been previously reported by Chandler et al. (2011), there appears to be atrial myocardium sandwiched between the two structures. 3.4) Discussion The extent of the SN measured in this study is comparable to what has been observed in many previous studies (Matsuyama et al., 2004; Sanchez-Quintana et al., 2005). It is however, smaller than the size observed by Chandler et al. (2011). In this study we were also unable to trace the superior extent of the paranodal area past the midpoint of the SN. This differs from the findings of Chandler et al. (2011) who traced both structures to the same level superiorly. There are a number of possible factors for these significant differences in structures seen. It has been observed that there can be significant variation in the length of sinus node in the human heart, for example Sanchez-Quintana et al. (2005) recorded lengths ranging from 8 mm to 21.5 mm, and that the length measured was not proportional to heart size or the length of the crista terminalis. It is therefore not unexpected that the morphology of the paranodal area appears likely to be highly variable from heart to heart. Despite the differences seen between the two reconstructions, the close proximity of the two structures was consistent. Variations in the overall size of the structures between studies may also be due to the tissue being processed differently. The right atrium is an elastic structure and tissue stretch prior to sectioning would lead to variability in the size of the structures. In this study there were areas where the SN myocytes appeared to be projecting into the atrial myocardium from the head and body regions of the SN (Fig 3.5). These extensions could be potential sites for exit pathways from the SN (as discussed on page 25). The presence of such pathways has been debated for many years. Optical 76 mapping of the SN appears to suggest that there are distinct sites of breakthrough for activation of the atrial myocardium by the SN (Fedorov et al., 2010). In the study by Sanchez-Quintana et al. (2005) SN extensions were found to project from the head, body and tail regions of the SN into the atrial myocardium. In the study by Chandler et al.(2011) the paranodal area was shown to extend past the inferior extent of the SN, however they were unable to trace the full extent of the paranodal area towards the inferior vena cava. In this study we have been able to show that the paranodal area continues to extend inferiorly along the crista terminalis, and reaches to within 5 mm of the end of the crista terminalis. A functional role for the extensive paranodal area has yet to be determined. The paranodal area is composed of a loosely packed combination of nodal-like and atriallike myocytes, and in particular is an area that has low Kir2.1 (inwardly-rectifying K+ channel), Cx40 and Cx43 expression (Chandler et al., 2009; Chandler et al., 2011). There is some potential for it to act as a pacemaker and many tachycardias in man have also been observed to originate from the region of the crista terminalis. Given that the paranodal area occupies a large portion of the crista terminalis, the arrhythmias may in fact be originating from the paranodal area (Figs 1.5 and 3.4). The leading pacemaker site is known not to be static and a pacemaker shift can occur, with activation observed along most of the length of the crista terminalis (Boineau et al., 1988). This 3D reconstruction of the human SN and complete paranodal area has the potential to be used for computer modelling studies. It could be used in order to help determine a functional role for the paranodal area which has not been possible previously as only partial reconstructions had been created. It could also be used to model the potential functional importance of specific exit pathways from the SN. 77 3.5) Limitations and future work Fibre orientation is known to be an important factor in action potential propagation through tissue (Ho et al., 2002). However, it was not possible to trace fibre orientation from the CT or histology in this study and so this information could not be incorporated into this reconstruction. In the future a contrast medium could be used during the CT scanning in order to obtain this information as previously shown by Stephenson et al. (2012). 78 CHAPTER 4) 3D ANATOMICAL RECONSTRUCTION OF THE COMPLETE CARDIAC CONDUCTION SYSTEM OF THE HUMAN HEART 4.1) Introduction Detailed 3D anatomical reconstructions of the whole human heart have previously been created based on MRI and CT data (Guo et al., 2005; Deng et al., 2012; ). These models allow the identification of large anatomical features of the heart such as atrial and ventricular trabeculations, the crista terminalis, atrioventricular valves and blood vessels. Currently however, it is not possible to identify the components of the cardiac conduction system (CCS) using these techniques alone. 3D reconstructions of individual components of the CCS have also been created based on information obtained from using histological and immunohistochemical techniques. These models are useful for looking at the detail of the structure of interest, for example the SN, but the extended role played in the CCS and the role of cardiac pacemaking cannot be assessed. 3D models of this nature are frequently used in computer simulation studies into arrhythmias. Combined models including detailed cardiac anatomy and the major components of the CCS conduction system are advantageous as they enable arrhythmogenic effects on the whole heart to be observed. Aim The aim of this study was to create a 3D reconstruction of the CCS of the whole human heart. The reconstruction would contain all of the major components of the CCS and demonstrate their morphologies and locations relative to each other. 4.2) Materials and methods 4.2.1) Whole human heart A whole human heart was obtained from Professor Paul Iaizzo from the University of Minnesota, USA. The heart was formalin fixed. 79 4.2.2) X-Ray Computed Tomography CT scanning was carried out using a Nikon Metris Custom Bay at the Manchester XRay Imaging Facility, University of Manchester. The sample was cleared of fixative by washing in distilled water and then immobilized in a holder to ensure there would be no movement during the imaging process. The scan was acquired using a copper filter with X-ray conditions of 132kV and 170µA. 2500 projections with an exposure time of 2s were collected over 360o. The whole human heart was scanned at a resolution of 85.8 µm. 4.2.3) Tissue preparation After CT scanning the whole heart was filled with chilled 2% carboxymethyl cellulose paste. It was then frozen in a mixture of hexane and solid carbon dioxide for 60 minutes. Sectioning was performed using a Leica CM3600 XP cryomacrotome (Leica Microsystems) at a thickness of 50 µm onto adhesive collection tape and allowed to air dry at room temperature. Six serial sections were collected and then 500 µm was discarded (forming each level) until the whole preparation had been sectioned. 950 sections were collected in total. To identify the CCS components Masson’s trichrome histology was performed on the tissue sections (Figs. 4.1 and 4.2; see section 3.2.4). On adjacent sections to those selected for histology immunohistochemistry was performed. VECTASTAIN Elite ABC kit (Vector Laboratories) was used. A similar initial protocol was used as for immunofluorescence staining (see section 3.2.5). Sections were initially treated with 0.3% H2O2 in methanol to block endogenous peroxidase activity followed by three 10 min washes in distilled water. The standard protocol was then followed up to incubation with the secondary antibody. The sections were then incubated in a biotinylated secondary antibody for 2 hours followed by three 10 min washes in PBS. The ABC (avidin and biotinylated horseradish peroxidase macromolecular complex) reagent is then applied for 60 minutes followed by three 10 min PBS washes. DAB (diaminobenzidine) solution is used to develop the signal. The sections were exposed using DAB for between two and five minutes. Anti-rabbit Cx43 and HCN4 IgG antibodies at concentrations of 1:50 were tested using this method on the tissue sections. It was not possible to detect any positive labelling of these proteins in these tissue sections. It is likely that the processing of the tissue prior to the immunohistochemistry being performed has damaged the proteins meaning they cannot be detected. Histology sections were imaged using a flatbed scanner. 80 Figure 4.1. Histology on whole human heart sections. A. Histology on whole human heart section. B. High magnification of sinus node region. C. High magnification of AVN region. 81 Figure 4.2. Histology on serial whole human heart sections. Sections shown from posterior (1) to anterior (12). 82 4.2.4) 3D anatomical reconstruction The CT images could not be used for the anatomical reconstruction as the chambers of the heart became compressed during the scanning process and so did not correspond well with the histological images (Fig. A2). This was due to the thin walls of the chambers. In the future it would be advantageous to fill the chambers with an X-ray inert substance such as agarose gel in order to support the structure of the chambers. 114 histological images were used for the reconstruction. The same procedure was followed as for the reconstruction of the isolated SN (see section 3.2.6). 4.3) Results I was able to create a 3D anatomical reconstruction of the whole human heart based on tissue sections of the intact heart (Fig. 4.3, Movie 4.1). The SN, retroaortic node, inferior nodal extensions, components of the AVN (compact node, penetrating bundle and nonbranching bundle) and the ventricular portion of the CCS (branching bundle and the left and right bundle branches) were identified and reconstructed. An accurate reconstruction was also made of the atrial and ventricular myocardium, the aorta and the aortic valve leaflets (Figs. 4.4 - 4.7). The size of the reconstructed structures is given in Table 4.1. The SN node is located in the intercaval region of the right atrium in close proximity to the crista terminalis. In this whole heart reconstruction the SN was a long crescent shaped structure approximately 2 cm in length from head to tail. This is comparable to the size found in isolated sinus node reconstructions. The retroaortic node was a cylindrical structure a cylindrical structure found in close proximity to the aorta. Two inferior nodal extensions were found in this heart. They originate in the posterior of the heart in the base of the right atrium. They unite and form the compact AVN. This compact node extends for approximately 5 mm towards the anterior of the heart. The penetrating bundle forms the sole conduction pathway through the central fibrous body linking the atrial and ventricular components. The penetrating bundle extends approximately 5 mm towards the apex of the heart. The non-branching His bundle is ~8.5 mm long and was situated within the ventricular septum. In this heart there appeared to be an extensive His bundle running within the ventricular septum for ~2 cm, from which the left and right bundle branches arise. 83 Figure 4.3. Whole heart anatomical reconstruction. Top: anterior view; Middle: medial view; Bottom: posterior view. 84 SN Aortic valve leaflets INE His Branching bundle LBB RBB Figure 4.4. Anterior view of 3D anatomical reconstruction of human CCS. A. Transparent working myocardium to show position of CCS and aorta. B. CCS reconstruction with working myocardium removed. INE: inferior nodal extension; LBB: left bundle branch; RBB: right bundle branch; SN: sinus node. 85 SN RAN CN Aortic valve leaflets Aortic valve leaflets INE His PB LBB Branching bundle RBB Figure 4.5. Medial view of 3D anatomical reconstruction of human CCS. A. Transparent working myocardium to show position of CCS and aorta. B. CCS reconstruction with working myocardium removed. CN: compact node; INE: inferior nodal extension; LBB: left bundle branch; PB: penetrating bundle; RAN: retroaortic node; RBB: right bundle branch; SN: sinus node. 86 SN Aortic valve leaflets CN Aortic valve leaflets RAN INE His LBB Branching bundle RBB Figure 4.6. Posterior view of 3D anatomical reconstruction of human CCS. A. Transparent working myocardium to show position of CCS and aorta. B. CCS reconstruction with working myocardium removed. CN: compact node; INE: inferior nodal extension; LBB: left bundle branch; RAN: retroaortic node; RBB: right bundle branch; SN: sinus node. 87 INE CN PB His Figure 4.7. CCS reconstruction with atria removed. Whole heart reconstuction digitally sectioned at level of the atrioventriuclar junction. Working myocardium is transparent. Morphololgy of AVN visible in right panel. CN: Compact node; INE: inferior nodal extension; PB: penetrating bundle. 88 Material Heart Sinus node Retroaortic node Inferior nodal extensions Compact node Penetrating bundle Non branching bundle Branching bundle Left bundle branch Right bundle branch Aorta Aortic valve leaflets Total Triangles 1723406 2938 2290 2870 1670 700 2564 9768 17750 9224 126590 14528 1914298 Volume 4262363 1201 771 887 743 89 1319 5063 8588 4784 71521 5729 4363058 Table 4.1. Whole heart model statistics. The number of triangles used to form the surface of each structure and the final volume of each tissue is given. 89 The aortic valve contained three leaflets. These valve leaflets are the left and right coronary leaflets and the more posteriorly located non-coronary leaflet. The closest structure to the basal attachment of the non-coronary cusp was the His bundle which was located 6 mm away (Fig. 4.8). 4.4) Discussion To the best of my knowledge this is the first 3D reconstruction of the CCS of the human heart to be created from a single intact heart. This model was created as a proof of principle that it was possible to section the intact human heart and then create an anatomical reconstruction of the CCS. A reconstruction created in this way has many advantages over previous reconstructions created by merging individually reconstructed components from differing sources into a single model. I was able to create an accurate representation of how the location of all of the structures relate to each other in situ. In many models and text books the CCS is dramatically oversimplified. A good knowledge of the location of the components of the CCS and their positioning relative to each other is vital in order to understand the functioning of the heart. The method used for this reconstruction also has the advantage that is shows the location of the components within the entire heart, without having to incorporate the information into geometry obtained from a separate heart or a stylized anatomical reconstruction. This information is important for clinicians to understand when planning treatments, such as catheter ablation or valve replacement, which have the potential to damage tissues close to the areas being treated. This anatomical information is also critical in order to be useful for accurate computer simulations of cardiac activity to be carried out. One of the disadvantages of this technique is that some fine detail of each individual structure may be lost due to the large size of the tissue sample. 4.4.1) Proximity of aortic valve to components The 3D reconstruction clearly shows the close proximity of the atrioventricular components of the CCS and the aortic valve. This is particularly important to consider when aortic valve replacement 90 procedures are being planned. Figure 4.8. Coronal slice through 3D reconstruction at level of aortic valve. 3D distance form non-coronary-cusp of aortic valve to His bundle shown. A Medtronic CoreValve is shown to give an indication of the positioning of aortic valve implants in relation to the CCS. Skirt length of CoreValve implant shown to indicate potential depth of implant in the left ventricular outflow tract. 91 It is relatively common for patients undergoing aortic valve replacement to develop arrhythmogenic complications and require additional treatment, such as the fitting of an artificial pacemaker (Dawkins et al., 2008). Aortic valve replacement is becoming a more commonly performed procedure with an increase from 7396 in 2004-2005 to 9333 in 2008-2009 in the UK alone (Dunning et al., 2011). This is partially down to an aging population, with an increase in the prevalence of valvular disease increasing with age. 1.3% of the population aged between 65 and 74 will have aortic stenosis, and by age 75-84 this will rise to 2.4% (Stewart et al., 1997). If aortic stenosis is left untreated it has a mortality rate of almost 80% after 3 years (Schwarz et al., 1982) demonstrating the importance of early intervention in treatment of this condition. Aortic stenosis is a disease of the aortic valve leaflets. It involves a calcification of the valve leaflets causing them to stiffen, therefore preventing them from function properly. The onset of calcification can be caused by rheumatic fever (Carabello, 2013) but now more commonly is seen as a disease of aging thought to be caused by inflammation due to hemodynamic stress (Carabello, 2013). If the aortic valve is not functioning properly it can lead to angina, low ejection fraction, syncope, dyspnea, and congestive heart failure (Schwarz et al., 1982). Aortic valve replacement can be split into surgical and non-surgical categories. Surgical replacement involves open heart surgery, the dissection of the diseased valve and the insertion of either a replacement mechanical or biological valve, held in place by sutures. There are obvious risks associated with this surgery as it requires a general anaesthetic and the opening of the chest wall and is unsuitable for many patients, particularly the elderly, which is an increasing problem with the rapidly aging population meaning that the age of patients requiring treatment is increasing. It is however considered as the gold standard for the treatment of aortic valve stenosis and accounts for around 90% of aortic valve replacements. Non-surgical replacement is performed using transcatheter techniques, so called transcatheter aortic valve implantation or replacement (TAVI/TAVR), and make up around 10% of aortic valve replacements. A catheter is inserted either into the femoral artery or through the chest wall and into the aorta. The native valve leaflets are compressed destroyed by balloon valvuloplasty. The valve implant is then positioned with the aid of fluoroscopy and is expanded in order to hold it in place. The two most commonly used aortic valve implants used in the TAVI procedure are the Medtronic CoreValve and the EdwardsSAPIEN. 92 Both consist of a metal cage in which is held bovine or porcine leaflets. These types of valve implants are not physically fixed in place by sutures, but are instead held in position solely by the stretching of the valve into the native aortic annulus. TAVI is a far less invasive procedure and has the potential to be use under conscious sedation. This makes its use potentially suitable for patients considered to be too high risk for surgical treatment, such as patients with comorbidities to aortic valve stenosis and the very elderly who may not be able to tolerate a general anaesthetic. There are risks with the TAVI procedure and there is evidence of increased chance of stroke, a lower one-year survival rate, and potential for the valve to become dislodged. Refinements in aortic valve replacement procedures with less invasive implantation techniques such as TAVI have meant that more patients are now suitable candidates for valve replacement and increasing numbers of aortic valve replacements are being performed. The average age of patients undergoing surgical replacement increased from 68.8 to 70.2 between 2004 and 2009, and an increase in the proportion of highrisk patients operated on increased from 24.6% to 27.7% over the same period (Dunning et al., 2011). Surgical aortic valve replacement remains the gold standard but TAVI is increasingly being performed on older and more complex patients with co-morbidities, who would not be suitable for surgical replacement, or for whom surgery is considered a high risk. Aortic valve replacement has been associated with a number of CCS defects. In particular there is evidence of new onset AVN block and left bundle branch block (Fig. 4.9) (Baan et al., 2010). Histology performed on sections from hearts from patients that had undergone surgical valve replacement showed traumatic haemorrhages in the region of the atrioventricular bundle and also areas where the atrioventricular bundle has been severed (Fukuda et al., 1976). 93 Figure 4.9. 12-lead ECG recording from patient undergoing TAVI procedure. Top. Prior to the TAVI procedure normal activation was found. Bottom. Complete left bundle branch block pattern post-TAVI. From van Dam et al. (2014) 94 The use of TAVI and the removal of the use of sutures may have been expected to be associated with fewer conduction abnormalities being caused, however even with TAVI conduction abnormalities are a common side effect. Approximately one third of patients undergoing TAVI will require the fitting of a permanent pacemaker (Khawaja et al., 2011). An examination of a heart after the use of the EdwardsSAPIEN implant revealed a haemorrhage in the region of the His bundle (Moreno et al., 2009). The positioning of the valve prosthesis is an important factor in determining whether a patient undergoing TAVI is likely to require the fitting of a pacemaker. When the implant is positioned in an infra-annular position, whereby the implant is situated below the native valve there is an increase in cardiac conduction defects, with significantly higher incidences of left bundle branch block in patients with an average implantation depth of 10.3 mm below the basal attachment of the noncoronary leaflet than those with an implantation depth of 5.5 mm (Piazza et al., 2008). This observation is not unexpected when the 3D model is examined. The His bundle is located just 6 mm from the aortic valve within the left ventricular outflow tract and so is susceptible to being damaged. The bundle branch is located approximately 12 mm from the aortic valve and so the lower the implant is positioned in the outflow tract the increased likelihood that there will be damage to the left bundle branch and subsequent conduction abnormalities. 4.4.2) Uses of the anatomical model This 3D computer anatomical model has many potential clinical and educational uses. In relation to aortic valve replacement this model may be useful in helping improve the design of the implants in order for them to have less of an impact on the CCS. The knowledge of the proximity of the CCS to the aortic valve could also help in the setting of implantation depth limits for the prosthetic valves as the implantation depth can vary widely and is known to be an important factor associated with subsequent conduction abnormalities. Computer simulations can be performed with this model to determine potential arrhythmias that may arise due to damage of specific conduction system components at specific locations. 95 These improvements to the TAVI procedure could lead to fewer procedural complications for the patients providing them with a better outcome. This anatomical model is also a valuable educational tool as it enables the proximity of and the interactions between the CCS to be more easily appreciated. This information is invaluable to people at all levels from clinicians to students. A detailed 3D anatomical reconstruction of the human heart and conduction system created from an intact heart could improve computer modelling of cardiac function as it includes the CCS components within their native atrial and ventricular locations, rather than being incorporated into geometries from different hearts, which can potentially lead to some problems if the geometries do not match perfectly, and this ensures that action potential propagation through the heart can be modelled more accurately. This anatomical model is currently being used in computer simulations into the conduction abnormalities caused by damage to the atrioventricular conduction components (Fig. A3). 4.5) Limitations and future work This piece of work was a proof of principle study, in order to show that it was possible to section the complete human heart and perform imaging techniques on the tissue sections that would allow the cardiac conduction system to be delineated. There are limitations with this reconstruction. Only one heart was reconstructed which means that whilst the general anatomy observed in this model is similar to previously reported, some care needs to be taken into reading conclusions from measurements made and interpreting interactions between the structures. It was also not possible with the current approach to reconstruct the Purkinje fibre network as the imaging resolutions of the histology and the CT scan were not high enough so as to be able to visualize them. I have performed a scan on a second human heart with a I2KI contrast medium. This scan will enable a far more detailed model, potentially including the Purkinje fibres and also fibre orientation as previously shown by Stephenson et al. (2012) to be created in the future. It would also be possible to segment out additional structures 96 and tissues. It would also be useful to create further reconstructions of healthy hearts to confirm the proximity of structures, create reconstructions of diseased hearts to see what anatomical changes occur to the CCS, hearts of different ages and also hearts that undergone the TAVI procedure to see if there are structural changes to the CCS. 97 CHAPTER 5) INVESTIGATING THE EXPRESSION OF THE LARGE CONDUCTANCE KCA1.1 CHANNEL IN THE HUMAN SINUS NODE 5.1) Introduction The KCa1.1channel is a member of the BK (big potassium) family of large conductance calcium-activated potassium channels. The BK channels are activated by increased intracellular Ca2+ and also by depolarizing membrane potentials (Bentzen et al., 2009) and have been shown to contribute to membrane repolarization and also affect Ca2+ handling in certain cell types due to their effects of the voltagegated Ca2+ channels (Turner and Zamponi, 2014). The BK channels have been linked to a number of physiological processes largely related to the nervous system, such as neurotransmitter release (Martire et al., 2010) and spontaneous activity in the suprachiasmatic nucleus in the hypothalamus (Meredith et al., 2006). They have also more recently been identified within the mitochondria of cardiomyocytes where they are associated with protection from ischemia (Xu et al., 2002), and also within smooth muscle cells within blood vessels, leading to effects on blood pressure (Brenner et al., 2000). Expression of the BK channels within the heart is thought to be low (Harrell et al., 2007) but a study involving the use of the BK channel inhibitors, paxilline and lolitrem, has found them to cause a reduction in heart rate in mice and rats whilst not affecting blood pressure (Imlach et al., 2010). This effect on heart rate was not observed in mice where the KCNMA1 gene which encodes the KCa1.1 channel was knocked out, suggesting that this channel in particular could play a role in cardiac pacemaking (Imlach et al., 2010) (Fig 5.1). The BK channels are composed of 4 pore forming α subunits, each consisting of 7 transmembrane domains with a Ca2+ bowl close to the C-terminus (Ghatta et al., 2006), and regulatory β subunits. The opening of the BK channels can lead to a hyperpolarization of the membrane potential (Carvalho-de-Souza et al., 2013), which may be expected to reduce cell excitability, and potentially act as part of a negative-feedback mechanism. 98 Figure 5.1. The effect of paxilline and iberiotoxin on heart rate in the isolated rat heart. A. 1 µM paxilline (Pax) added after 40 min and infused for 10 min, following a control infusion of Krebs-Henseleit perfusion fluid (n=5). B. Heart rate before, during and after infusion at time zero of different concentrations of paxilline (1, 5, 10 µM). C. Heart rate of isolated, perfused rat hearts infused with 0.23 µM iberiotoxin (IbTX) following a control infusion of Krebs-Henseleit fluid (n=5). D. Heart rate before, during and after infusion of iberiotoxin (n=5). All data are mean ± S.E.M. *** P<0.001. From (Imlach et al., 2010) 99 Aim The aim of this study was to determine whether there is a higher expression of KCa1.1protein in the SN compared to the paranodal area and atrial muscle, therefore, determining whether it could potentially be used as a marker for the human SN. Differential expression was also examined at the mRNA level for the KCNMA1 gene within these tissues. 5.2) Materials and methods 5.2.1) Molecular investigation Tissue sampling Frozen human SN tissue sections from 4 hearts (see Table 2.1) were selected at approximately 500µm intervals and used for Masson trichrome histology, and immunohistochemistry using HCN4 and Cx43 antibodies, in order to determine the precise location of the SN and paranodal area in the sections (see section 3.2.6). The remaining sections were subjected to a quick hematoxylin and eosin staining protocol. Sections were placed in RNase free water for 30 sec, then Mayer’s hematoxylin (Sigma Aldrich) for 1 min. They were then washed twice for 30 sec in 100% ethanol. In order to preserve RNA quality alcoholic solutions were used. Sections were stained with alcoholic Eosin Y solution (Sigma Aldrich) for 1 min and washed 3 times for 30 sec in 100% ethanol. Sections were kept in 100% ethanol until used for dissection. Dissection was carried out using a sterile fine needle to lift tissue from slide. Samples were taken from SN, paranodal area and right atrium from approximately 10 sections per heart. Dissected samples were collected in 600 µl lysis buffer solution containing β-mercaptoethanol from the PureLink RNA Mini isolation kit (Applied Biosystems). RNA isolation RNA isolation was performed using the PureLink RNA Mini isolation kit (Applied Biosystems) and all solutions prepared in accordance with the manufacturer’s instructions. The dissected tissue was homogenized in the 600µl lysis buffer containing β-mercaptoethanol for 90 seconds using a homogenizer (Ika-Werke). Samples were centrifuged at 26000g for 5 minutes in order to remove cell debris that 100 could interfere with the RNA isolation. After centrifugation the supernatant is retrieved and is thoroughly mixed with an equal volume of 70% ethanol. This mixture is then loaded into a PureLink spin cartridge and centrifuged at 12000g for 15 seconds, after which the flow through was discarded. 350µl wash buffer I is added to the columns and they are then centrifuged at 12000g for 15 seconds. The samples were then treated with on-column PureLink DNase (Applied Biosystems). The DNase was prepared using 8µl 10X DNase I reaction buffer, 10µl DNase [3U/µl], and 62µl RNase-free water per sample. 80µl of this DNase mixture was added to the centre of the spin cartridge membrane and incubated at room temperature for 15 minutes. 350µl wash buffer I is added to the columns and they are then centrifuged at 12000g for 15 seconds. 500µl wash buffer II is added to the samples and spun at 12000g for 15 seconds. This was step is repeated and the flowthrough is discarded. The spin cartridge is then further centrifuged for 1 minute in order to dry out the membrane. RNase-free water is used for the RNA elution. 30µl RNase-free water is added to the centre of the cartridge membrane and incubated at room temperature for 1 minute. The spin cartridge in centrifuged in a recovery tube at 12000g for 2 minutes and the RNA containing elute recovered. The yield of total RNA was quantified using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific). Reverse transcription High Capacity RNA-to-cDNA Master Mix (Life Technologies) was used for reverse transcription. The master mix contains all of the components required for a cDNA synthesis reaction including reverse transcriptase, RNase Inhibitor Protein, dNTPs, MgCl2 and primers ((a blend of random primers and oligo(dT)). 170 ng total RNA in 16 µl water was added to 4 µl master mix. The reaction was run for 5 min at 25oC (primer annealing), 30 min at 42oC (reverse transcription), 5 min at 85oC (denature enzyme) and finally held at 4oC using a Veriti Termal Cycler (Applied Biosystems). Samples were stored at -80oC until use. 101 Primers Custom primers designed against the C terminus region and the pore forming region of the KCNMA1 gene and a commercially available KCNMA1 QuantiTect primer assay (Qiagen) were used (Table 5.1 A+B). QuantiTect primers against TBX3 and GJA1 (Cx43) were used to characterise the sampled tissues, and confirm SN tissue has been successfully collected (Table 5.1 B). Custom designed primers against the 28s housekeeper gene were used (Table 5.1A). qPCR reaction setup When QuantiTect primers were used each 10 µl reaction for qPCR contained 1 µl cDNA, 1 µl QuantiTect primer (Qiagen), 5 µl Power SYBR green master mix (Applied Biosystems), 3 µl water. When custom designed primers were used each 10 µl reaction for qPCR contained 1 µl cDNA, 0.4 µl forward primer, 0.4 µl reverse primer, 5 µl Power SYBR green master mix (Applied Biosystems), 3.2 µl water. Forward and reverse primers were used at concentrations of 50 nm, 300 nm and 900 nm. qPCR was performed using ABI 7900HT apparatus (Applied Biosystems). Samples were heated to 50oC for 2 min, followed by 10 min at 95oC. The samples then underwent 40 cycles of 95oC for 15 sec and 60oC for 1min. Fluorescence measurements for each reaction are made every cycle. Data analysis ΔCt values were calculated by normalising Ct values for each target gene to those obtained for the 28s ribosomal gene for each sample. Outliers were determined using a robust modified z-score method based on the median of absolute deviation (Iglewicz and Hoaglin, 1993). One-way ANOVA was used to determine significant differences in expression between the regions sampled. A Holm-Sidak post-hoc test was used to test for significant differences between tissues. P<0.05 was taken to be significant. 102 A Target mRNA Sequence 28S Forward Reverse - GTTGTTGCCATGGTAATCCTGCTCAGTACG TCTGACTTAGAGGCGTTCAGTCATAATCCC KCNMA 1 (C Terminus) Forward Reverse - TGTGCACCCAAGGAGATAGA GATGTTGAGTGACGCCAAGA KCNMA 1 (Pore Region) Forward Reverse - CCTGATCCTTGCCAACAAGT AGCTCGGGATGTTTAGCAGA B Target mRNA QuantiTect assay KCNMA1 TBX3 GJA1 QT00024157 QT00022484 QT00012684 Table 5.1. qPCR primers used. A. Custom designed primers for 28S housekeeper gene and KCNMA1. B. Qiagen QuantiTect assays. 103 5.2.2) Protein investigation KCa immunohistochemistry Immunohistochemistry using antibodies against the KCa1.1 protein was used to assess the protein expression within the SN compared to the paranodal area and atrial myocardium. SN tissue sections were selected from 4 hearts (Table 2.1). Three rabbit polyclonal IgG antibodies (all Alomone Labs) targeted to different regions of the protein were initially trialled. Two of the antibodies were targeted at intracellular regions of the protein and one an extracellular portion (Table 2.2). The protocol used was the same a previously described (section 3.2.5). The optimal antibody concentration was determined using a range of concentrations from 1:50 to 1:800. In order to determine the location of the protein within the cardiac tissue more accurately the sections stained with KCa1.1 antibodies were double labelled with guinea-pig polyclonal anti-vimentin (Progen), mouse monoclonal anti-Cx43 (Millipore), Smooth muscle α actin (Sigma Aldrich) and mouse monoclonal antiRYR IgG (Thermo Scientific) antibodies (see Table 2.2). Signal intensity measurement The signal intensity of immunofluorescence for each antibody was measured using Volocity software (Improvision, UK). One-way ANOVA was used to determine significant differences in expression between the regions sampled. A Holm-Sidak post-hoc test was used to test for significant differences between tissues. P<0.05 was taken to be significant. 5.3) Results 5.3.1) qPCR Both of the custom designed primer sets failed to amplify, whilst the QuantiTect assays amplified successfully (Fig 2.4A). A melt-curve was used to check for the amplification of a single product. A single product was confirmed by the presence of a single dissociation peak (Fig 2.4B). Only data obtained using the QuantiTect primers was used for subsequent analysis. No significant differences were found between the atrial muscle, paranodal area and sinus node for TBX3, GJA1 or 104 KCNMA1. Expression of TBX3 in the SN showed a tendency to be higher than in the other tissues (Fig 5.2). GJA1 expression was lowest in the sinus node, and it appeared highest in the paranodal area (Fig 5.2). The SN showed a tendency to have higher expression of KCNMA1 than the paranodal area or atrial muscle (Fig 5.2). 5.3.2) KCa protein investigation The antibody that provided the clearest and most consistent staining for KCa1.1 was targeted at residues 1097-1196 in the intracellular C-terminus (APC021). The staining appeared to be both within the cell membrane and intracellular (Fig 5.3 top). The antibody targeted against the extracellular 1st loop (APC151) also provided cell membrane and intracellular staining (Fig 5.3 bottom). The signal produced by the second intracellular antibody targeted at residues 1184-1200 (APC107) appeared to be solely intracellular, with no apparent surface membrane labelling (Fig 5.3 middle). Strong labelling within the fibroblasts located between cardiomyocytes was observed (Fig 5.3 top). Negative control experiments using APC021 and APC151 antibodies showed a lack of staining, although some auto fluorescence from the tissue was detectable (Figs. 5.4 and 5.5) 5.3.2.1) Dilution The optimum concentration for APC021 and APC151 antibodies was determined to be 1:50 (Figs 5.4 and 5.5). Below this concentration the labelling achieved was not strong enough to visualize the protein localisation. I used these antibodies at this 1:50 concentration for the subsequent double labelling experiments. 5.3.2.2) Localisation Double labelling using the KCa1.1 antibodies with ryanodine receptor 2 (Ryr2), Cx43, Smooth muscle α actin and vimentin antibodies helped clarify the cellular localisation of the KCa1.1 signal. Ryr2 labelling is found within the sarcoplasmic reticulum, and there appeared to be large areas where the signal overlapped, suggesting that the proteins are present in the same cellular locations (Fig 5.3 right). Cx43 is located at the intercalated disk and there was co-localisation was observed (Fig 5.3 left). Vimentin is a fibroblast marker within the heart, and there appeared to 105 be some co-localization suggesting that the KCa1.1 protein may also be present in fibroblasts as well as cardiomyocytes (Fig. A4). Overall the expression of KCa1.1 was found to be present at both the cell surface membrane and also within the sarcoplasmic reticulum membrane of cardiomyocytes. It would be necessary to look at isolated single cells to better determine the relative expression in each cell type. 106 Figure 5.2. mRNA expression in the SN, PN and AM. mRNA expression normalised to 28s housekeeper gene of KCNMA1 (top), TBX3 (middle) and GJA1 (bottom). 107 Figure 5.3. Localisation of KCa1.1 protein. Immunohistochemistry using various KCa1.1 antibodies with Cx43 and Ryr2 antibodies. Double labelling of: Top left: KCa1.1 (APC021; green) and Cx43 (red). Top right: KCa1.1 (APC021; green) and Ryr2 (red). Middle left: KCa1.1 (APC107; green) and Cx43 (red). Middle right: KCa1.1 (APC107; green) and Ryr2 (red). Bottom left: KCa1.1 (APC151; green) and Cx43 (red). Bottom right: KCa1.1 (APC151; green) and Ryr2 (red). 108 Figure 5.4. APC012 KCa1.1 antibody concentration optimisation. Immunohistochemistry on human right ventricle using various concentrations of KCa1.1 antibody to determine optimal concentration. Concentration used shown in bottom left of each image. 109 Figure 5.5. APC151 KCa1.1 antibody concentration optimisation. Immunohistochemistry on human right ventricle using various concentrations of KCa1.1 antibody to determine optimal concentration. Concentration used shown in bottom left of each image. 110 5.3.2.3) Signal Quantification Signal intensity of fluorescence images was measured over a scale of 1 to 255. Overall the KCa1.1 signal intensity observed was low with average values for all regions being below 40. No significant difference was observed in the KCa1.1 fluorescence between the atrial muscle, paranodal area and SN. However the sinus node did show a slight tendency towards a higher signal intensity than the other tissues suggesting KCa1.1 protein may be higher in the SN (Fig 5.6). 5.4) Discussion KCa1.1 is a member of the BK family of large conductance calcium-activated potassium channels that have wide ranging roles in many excitable cell types in the human. The results from this study, whilst not being conclusive, suggest that the human SN expresses the KCa1.1 channel at higher levels than the atrial working myocardium. The SN showed a tendency for higher KCa1.1 expression at both the mRNA and protein levels (Figs 5.2+5.6), and therefore it may have a role in human cardiac pacemaking and potentially in arrhythmogenesis. The study by Imlach et al. (2010) suggested for the first time that the KCa1.1 channel may have a functional role in cardiac pacemaking in rats and mice, and with the results from this study suggests that further investigation in to the role of KCa1.1 within the myocytes of the SN in humans is warranted and that they could be a potential target in the treatment of arrhythmias. Pacemaking within the heart is thought to be due to a combination of the membrane voltage clock and the calcium clock mechanisms (Fig 5.7). The membrane clock is associated with the funny current (If) that is carried by the HCN1 and 4 channels (Baruscotti et al., 2010). This creates a spontaneous diastolic depolarization of the membrane potential, leading to the opening of the T-type voltage-gated Ca2+ channels (Cav3.1-3.3), and rapid membrane depolarization. The calcium clock meanwhile is a mechanism associated with calcium release from the sarcoplasmic reticulum via the ryanodine receptor, which then activates membrane bound NCX leading to a depolarizing current due to the electrogenic exchange of Na+ and Ca2+. 111 Figure 5.6. KCa1.1 expression in SN, PN and AM quantification. Representative images of KCa1.1 expression in SN, PN and AM in the human heart. Graph shows signal intensity quantification. 112 Figure 5.7. Pacemaking mechanisms in the sinus node. A. Currents and corresponding ion channels and transporters involved in pacemaking in the sinus node. B. Typical sinus node action potential and contributing ion channels. From Dobrzynski et al. (2013). 113 Calcium also enters the cell via the L-type and T-type voltage gated Ca2+ channels to further drive the exchange of Na+ and Ca2+. Calcium is taken back into the sarcoplasmic reticulum from the cytosol by sarcoplasmic reticulum Ca2+-ATPase (SERCA). Functionally, there is evidence that the KCa1.1 channel forms complexes with Cav1 and Cav3 voltage-gated calcium channels in neuronal cell types, leading to the activation of KCa1.1 over a larger voltage range and contributing to a rapid repolarization of the cell membrane (Vandael et al., 2010; Rehak et al., 2013). If this was also true for cardiomyocytes within the SN then it could provide an important link into explaining how membrane and calcium clock pacemaking mechanisms are potentially connected. KCa1.1 could be involved in the repolarization phase of the cardiac action potential, leading to a shortening of the action potential duration. BK channel blockers have been shown to lead to an increased APD in rat and mouse chromaffin cells (Vandael et al., 2010). Therefore, the blocking of the KCa1.1 channel would potentially cause a reduction in heart rate due to this increased action potential duration, which may explain the observations of Imlach et al. (2010). A prolonged action potential duration is a known risk factor for the development of cardiac arrhythmias, such as early after depolarizations and so the KCa1.1 channel may be a potential target in their treatment. 5.5) Limitations This investigation was underpowered as it was performed using tissue obtained from only 4 hearts. In the future a larger number of hearts would need to be used in order to properly assess expression differences. The expression pattern of the KCa1.1 protein visualized by immunohistochemistry makes the signal intensity quantification less reliable. In order to overcome this, we would need to use a larger number of tissue sections and ideally a larger number of hearts. 114 CHAPTER 6) GENERAL DISCUSSION The creation of 3D models is an increasingly important field and they have many potentially valuable educational, research and clinical uses. For educational purposes it is invaluable to be able to visualise the location of the major features of the heart including the CCS. Many current representations of the CCS are inadequate as they are often over simplified. A computer model can be easily modified to suit the target audience with the addition or removal of specific structures. In 2D representations it is difficult to ascertain how structures are interrelated with each other, whilst the 3D model allows the complete structure to be explored. The 3D model is also important in the potential to reorientate the heart into an attitudinally correct orientation. This is particularly important as it is a regular criticism of many anatomical studies that they alter the orientation of the heart from that seen in the body which can lead to the confusing naming of structures and tracing of major structures. This would be an important for surgeons to appreciate how they may potentially affect critical components of the heart. Accurate reconstructions of cardiac anatomy and the conduction system are also useful to the research community. Computer models of action potentials for different regions of the heart are being widely developed (Noble and Rudy, 2001; Boyett et al., 2005). It is possible for these action potentials to be incorporated into the 3D geometry. Simulations can then be run to assess whether arrhythmias may arise under different conditions such as with the introduction of ectopic foci and altered conduction properties, and also how these arrhythmias may potentially be terminated, such as through ablation of certain regions. These simulations are also potentially important in the development of drugs as it is possible to alter the characteristics of ionic currents and assess the effect they have an activation patterns in the heart, whether they are able to reduce the susceptibility to arrhythmia production or whether there are any potentially pro arrhythmic side effects. There is also a potential clinical use for 3D computer models. Any surgery on the heart can have potentially damaging effects due to inadvertent damage to the CCS. This can often be due to the complex anatomy of the CCS not being fully understood and its close proximity to structures that require surgery. In aortic valve replacement procedures cardiac arrhythmias are a common complication and often require the 115 subsequent fitting of artificial pacemakers (Dawkins et al., 2008). It has been shown that it is often portions of the AVN and the bundle branches that are damaged (Baan et al., 2010; Laynez et al., 2011). 3D models could be used to improve the design of cardiac implants so as to minimise their impact of important conduction structures and also to help improve procedures so that the vulnerable components of the cardiac conduction system are avoided where possible. Computer models are becoming increasingly highly detailed with far more accurate 3D anatomical geometries and complex cell models meaning that their potential uses are expanding and their importance increasing rapidly. 116 REFERENCES Ambrosi, C. M., Moazami, N., Rollins, A. M., Efimov, I. R. (2009). Virtual histology of the human heart using optical coherence tomography. J.Biomed.Opt. 14: 054002. Anderson, R. H., Cook, A. C. (2007). The structure and components of the atrial chambers. Europace 9 Suppl 6: vi3-9. Anderson, R. H., Ho, S. Y. (1998). The architecture of the sinus node, the atrioventricular conduction axis, and the internodal atrial myocardium. J.Cardiovasc.Electrophysiol. 9: 1233-1248. Anderson, R. H., Yanni, J., Boyett, M. R., Chandler, N. J., Dobrzynski, H. (2009). The anatomy of the cardiac conduction system. Clin.Anat. 22: 99-113. Ansari, A., Ho, S. Y., Anderson, R. H. (1999). Distribution of the Purkinje fibres in the sheep heart. Anat Rec 254: 92-97. Aschoff, L. (1910). Referat uber die Herzstorungen in ihren Beziehungen zu den Spezifischen Muskelsystem des Herzens. Verh Dtsch Pathol Ges 14: 3-35. Aslanidi, O. V., Colman, M. A., Stott, J., Dobrzynski, H., Boyett, M. R., Holden, A. V., Zhang, H. (2011). 3D virtual human atria: A computational platform for studying clinical atrial fibrillation. Prog.Biophys.Mol.Biol. 107: 156-168. Atkinson, A. J., Logantha, S. J. R. J., Hao, G., Yanni, J., Fedorenko, O., Sinha, A., Gilbert, S. H., Benson, A. P., Buckley, D. L., Anderson, R. H., Boyett, M. R., Dobrzynski, H. (2013). Functional, Anatomical, and Molecular Investigation of the Cardiac Conduction System and Arrhythmogenic Atrioventricular Ring Tissue in the Rat Heart. J Am Heart Assoc 2. Baan, J., Yong, Z. Y., Koch, K. T., Henriques, J. P., Bouma, B. J., Vis, M. M., Cocchieri, R., Piek, J. J., de Mol, B. A. (2010). Factors associated with cardiac conduction disorders and permanent pacemaker implantation after percutaneous aortic valve implantation with the CoreValve prosthesis. Am Heart J 159: 497-503. Bancroft, J. D., Gamble, M. (2008). Theory and practice of histological techniques, Churchill Livingstone Elsevier. Baruscotti, M., Barbuti, A., Bucchi, A. (2010). The cardiac pacemaker current. J Mol Cell Cardiol 48: 55-64. Bentzen, B. H., Osadchii, O., Jespersen, T., Hansen, R. S., Olesen, S. P., Grunnet, M. (2009). Activation of big conductance Ca(2+)-activated K (+) channels (BK) protects the heart against ischemia-reperfusion injury. Pflugers Arch 457: 979-988. 117 Benvenuti, L. A., Aiello, V. D., Higuchi, M. L., Palomino, S. A. (1997). Immunohistochemical expression of atrial natriuretic peptide (ANP) in the conducting system and internodal atrial myocardium of human hearts. Acta Histochem. 99: 187-193. Billinton, N., Knight, A. W. (2001). Seeing the wood through the trees: a review of techniques for distinguishing green fluorescent protein from endogenous autofluorescence. Anal Biochem 291: 175-197. Biosyn.com. (2014). "TaqMan® vs. SYBR® Green Chemistries." Retrieved 20/06/2014, from http://www.biosyn.com/tew/taqman-vs-sybr-greenchemistries.aspx. Boenisch, T. (2005). Effect of heat-induced antigen retrieval following inconsistent formalin fixation. Appl Immunohistochem Mol Morphol 13: 283-286. Boineau, J. P., Canavan, T. E., Schuessler, R. B., Cain, M. E., Corr, P. B., Cox, J. L. (1988). Demonstration of a widely distributed atrial pacemaker complex in the human heart. Circulation 77: 1221-1237. Boyett, M. R., Li, J., Inada, S., Dobrzynski, H., Schneider, J. E., Holden, A. V., Zhang, H. (2005). Imaging the heart: computer 3-dimensional anatomic models of the heart. J Electrocardiol 38: 113-120. Breithardt, G., Breithardt, O. A. (2012). Left bundle branch block, an old-new entity. J Cardiovasc Transl Res 5: 107-116. Brenner, R., Perez, G. J., Bonev, A. D., Eckman, D. M., Kosek, J. C., Wiler, S. W., Patterson, A. J., Nelson, M. T., Aldrich, R. W. (2000). Vasoregulation by the beta1 subunit of the calcium-activated potassium channel. Nature 407: 870-876. Buchwalow, I. B., Böcker, W. (2010). Immunohistochemistry: Basics and Methods: Basics and Methods, Springer. Carabello, B. A. (2013). Introduction to aortic stenosis. Circ Res 113: 179-185. Carvalho-de-Souza, J. L., Varanda, W. A., Tostes, R. C., Chignalia, A. Z. (2013). BK Channels in Cardiovascular Diseases and Aging. Aging Dis 4: 38-49. Chandler, N., Aslanidi, O., Buckley, D., Inada, S., Birchall, S., Atkinson, A., Kirk, D., Monfredi, O., Molenaar, P., Anderson, R., Sharma, V., Sigg, D., Zhang, H., Boyett, M., Dobrzynski, H. (2011). Computer three-dimensional anatomical reconstruction of the human sinus node and a novel paranodal area. Anat.Rec.(Hoboken.) 294: 970-979. Chandler, N. J., Greener, I. D., Tellez, J. O., Inada, S., Musa, H., Molenaar, P., Difrancesco, D., Baruscotti, M., Longhi, R., Anderson, R. H., Billeter, R., Sharma, V., Sigg, D. C., Boyett, M. R., Dobrzynski, H. (2009). Molecular architecture of the 118 human sinus node: insights into the function of the cardiac pacemaker. Circulation 119: 1562-1575. Davies, J., Anderson, R. H., Anton, E. B. (1983). The conduction system of the heart, Butterworths. Dawkins, S., Hobson, A. R., Kalra, P. R., Tang, A. T., Monro, J. L., Dawkins, K. D. (2008). Permanent pacemaker implantation after isolated aortic valve replacement: incidence, indications, and predictors. Ann Thorac Surg 85: 108-112. Demoulin, J. C., Kulbertus, H. E. (1978). Histopathological correlates of sinoatrial disease. Br Heart J 40: 1384-1389. Denes, P., Wu, D., Dhingra, R. C., Chuquimia, R., Rosen, K. M. (1973). Demonstration of dual A-V nodal pathways in patients with paroxysmal supraventricular tachycardia. Circulation 48: 549-555. Deng, D., Jiao, P., Ye, X., Xia, L. (2012). An image-based model of the whole human heart with detailed anatomical structure and fiber orientation. Comput.Math.Methods Med. 2012: 891070. Dobrzynski, H., Anderson, R. H., Atkinson, A., Borbas, Z., D'Souza, A., Fraser, J. F., Inada, S., Logantha, S. J., Monfredi, O., Morris, G. M., Moorman, A. F., Nikolaidou, T., Schneider, H., Szuts, V., Temple, I. P., Yanni, J., Boyett, M. R. (2013). Structure, function and clinical relevance of the cardiac conduction system, including the atrioventricular ring and outflow tract tissues. Pharmacol Ther 139: 260-288. Dunning, J., Gao, H., Chambers, J., Moat, N., Murphy, G., Pagano, D., Ray, S., Roxburgh, J., Bridgewater, B. (2011). Aortic valve surgery: marked increases in volume and significant decreases in mechanical valve use-an analysis of 41,227 patients over 5 years from the Society for Cardiothoracic Surgery in Great Britain and Ireland National database. J Thorac Cardiovasc Surg 142: 776-782 e773. Fedorov, V. V., Glukhov, A. V., Chang, R., Kostecki, G., Aferol, H., Hucker, W. J., Wuskell, J. P., Loew, L. M., Schuessler, R. B., Moazami, N., Efimov, I. R. (2010). Optical mapping of the isolated coronary-perfused human sinus node. J.Am.Coll.Cardiol. 56: 1386-1394. Fukuda, T., Hawley, R. L., Edwards, J. E. (1976). Lesions of conduction tissue complicating aortic valvular replacement. Chest 69: 605-614. Gaborit, N., Le Bouter, S., Szuts, V., Varro, A., Escande, D., Nattel, S., Demolombe, S. (2007). Regional and tissue specific transcript signatures of ion channel genes in the non-diseased human heart. J Physiol 582: 675-693. Gaskell, W. H. (1900). The contraction of cardiac muscle. Textbook of Physiology. E. A. Schäfer. Edinburgh and London Young JPentland: 169-227. 119 Ghatta, S., Nimmagadda, D., Xu, X., O'Rourke, S. T. (2006). Large-conductance, calcium-activated potassium channels: structural and functional implications. Pharmacol Ther 110: 103-116. Gomes, J. A., Winters, S. L. (1987). The Origins of the Sinus Node Pacemaker Complex in Man - Demonstration of Dominant and Subsidiary Foci. Journal of the American College of Cardiology 9: 45-52. Greener, I. D., Monfredi, O., Inada, S., Chandler, N. J., Tellez, J. O., Atkinson, A., Taube, M. A., Billeter, R., Anderson, R. H., Efimov, I. R., Molenaar, P., Sigg, D. C., Sharma, V., Boyett, M. R., Dobrzynski, H. (2011). Molecular architecture of the human specialised atrioventricular conduction axis. J.Mol.Cell Cardiol. 50: 642-651. Grossman, J. I., Delman, A. J. (1969). Serial P wave changes in acute myocardial infarction. American Heart Journal 77: 336-341. Guo, Y. L., Heng, P. A., Zhang, S. X., Liu, Z. J., Tan, L. W., Li, Q. Y., Qiu, M. G., Li, K., Fan, H. Q., Wang, Y. S., Tang, Z. S. (2005). Thin sectional anatomy, threedimensional reconstruction and visualization of the heart from the Chinese Visible Human. Surg Radiol Anat 27: 113-118. Harrell, M. D., Harbi, S., Hoffman, J. F., Zavadil, J., Coetzee, W. A. (2007). Largescale analysis of ion channel gene expression in the mouse heart during perinatal development. Physiol Genomics 28: 273-283. Ho, S. Y., Anderson, R. H., Sanchez-Quintana, D. (2002). Atrial structure and fibres: morphologic bases of atrial conduction. Cardiovasc Res 54: 325-336. Ho, S. Y., Sanchez-Quintana, D. (2009). The importance of atrial structure and fibers. Clin Anat 22: 52-63. Hucker, W. J., Fedorov, V. V., Foyil, K. V., Moazami, N., Efimov, I. R. (2008a). Images in cardiovascular medicine. Optical mapping of the human atrioventricular junction. Circulation 117: 1474-1477. Hucker, W. J., McCain, M. L., Laughner, J. I., Iaizzo, P. A., Efimov, I. R. (2008b). Connexin 43 expression delineates two discrete pathways in the human atrioventricular junction. Anat.Rec.(Hoboken.) 291: 204-215. Iglewicz, B., Hoaglin, D. C. (1993). How to Detect and Handle Outliers, ASQC Quality Press. Imlach, W. L., Finch, S. C., Miller, J. H., Meredith, A. L., Dalziel, J. E. (2010). A role for BK channels in heart rate regulation in rodents. PLoS.One. 5: e8698. Inoue, S., Becker, A. E. (1998). Posterior extensions of the human compact atrioventricular node: a neglected anatomic feature of potential clinical significance. Circulation 97: 188-193. 120 Irisawa, H., Seyama, I. (1966). Configuration of P wave during mild exercise. American Heart Journal 71: 467-472. James, T. N. (2001). The internodal pathways of the human heart. Prog.Cardiovasc.Dis. 43: 495-535. James, T. N., Sherf, L., Fine, G., Morales, A. R. (1966). Comparative ultrastructure of the sinus node in man and dog. Circulation 34: 139-163. Jongsma, H. J., Wilders, R. (2000). Gap junctions in cardiovascular disease. Circ Res 86: 1193-1197. Kalbfleisch, S. J., Strickberger, S. A., Williamson, B., Vorperian, V. R., Man, C., Hummel, J. D., Langberg, J. J., Morady, F. (1994). Randomized comparison of anatomic and electrogram mapping approaches to ablation of the slow pathway of atrioventricular node reentrant tachycardia. J Am Coll Cardiol 23: 716-723. Kalman, J. M., Olgin, J. E., Karch, M. R., Hamdan, M., Lee, R. J., Lesh, M. D. (1998). "Cristal tachycardias": origin of right atrial tachycardias from the crista terminalis identified by intracardiac echocardiography. J Am Coll Cardiol 31: 451459. Keith, A., Flack, M. (1907). The Form and Nature of the Muscular Connections between the Primary Divisions of the Vertebrate Heart. J.Anat.Physiol 41: 172-189. Khawaja, M. Z., Rajani, R., Cook, A., Khavandi, A., Moynagh, A., Chowdhary, S., Spence, M. S., Brown, S., Khan, S. Q., Walker, N., Trivedi, U., Hutchinson, N., De Belder, A. J., Moat, N., Blackman, D. J., Levy, R. D., Manoharan, G., Roberts, D., Khogali, S. S., Crean, P., Brecker, S. J., Baumbach, A., Mullen, M., Laborde, J. C., Hildick-Smith, D. (2011). Permanent pacemaker insertion after CoreValve transcatheter aortic valve implantation: incidence and contributing factors (the UK CoreValve Collaborative). Circulation 123: 951-960. Kistler, P. M., Roberts-Thomson, K. C., Haqqani, H. M., Fynn, S. P., Singarayar, S., Vohra, J. K., Morton, J. B., Sparks, P. B., Kalman, J. M. (2006). P-wave morphology in focal atrial tachycardia: development of an algorithm to predict the anatomic site of origin. J Am Coll Cardiol 48: 1010-1017. Kurian, T., Ambrosi, C., Hucker, W., Fedorov, V. V., Efimov, I. R. (2010). Anatomy and electrophysiology of the human AV node. Pacing Clin.Electrophysiol. 33: 754762. Laynez, A., Ben-Dor, I., Hauville, C., Xue, Z., Satler, L. F., Kent, K. M., Pichard, A. D., Lindsay, J., Waksman, R. (2011). Frequency of cardiac conduction disturbances after balloon aortic valvuloplasty. Am J Cardiol 108: 1311-1315. Majno, G., Joris, I. (2004). Cells, Tissues, and Disease : Principles of General Pathology: Principles of General Pathology, Oxford University Press, USA. 121 Martire, M., Barrese, V., D'Amico, M., Iannotti, F. A., Pizzarelli, R., Samengo, I., Viggiano, D., Ruth, P., Cherubini, E., Taglialatela, M. (2010). Pre-synaptic BK channels selectively control glutamate versus GABA release from cortical and hippocampal nerve terminals. J Neurochem 115: 411-422. Massing, G. K., James, T. N. (1976). Anatomical configuration of the His bundle and bundle branches in the human heart. Circulation 53: 609-621. Matsuyama, T. A., Inoue, S., Kobayashi, Y., Sakai, T., Saito, T., Katagiri, T., Ota, H. (2004). Anatomical diversity and age-related histological changes in the human right atrial posterolateral wall. Europace. 6: 307-315. Meredith, A. L., Wiler, S. W., Miller, B. H., Takahashi, J. S., Fodor, A. A., Ruby, N. F., Aldrich, R. W. (2006). BK calcium-activated potassium channels regulate circadian behavioral rhythms and pacemaker output. Nat Neurosci 9: 1041-1049. Monckeberg, J. (1910). Beitrage zur normalen und pathologischen Anatomie des Herzens. Verh Dtsch Pathol Ges 14: 64-71. Moreno, R., Dobarro, D., Lopez de Sa, E., Prieto, M., Morales, C., Calvo Orbe, L., Moreno-Gomez, I., Filgueiras, D., Sanchez-Recalde, A., Galeote, G., JimenezValero, S., Lopez-Sendon, J. L. (2009). Cause of complete atrioventricular block after percutaneous aortic valve implantation: insights from a necropsy study. Circulation 120: e29-30. Noble, D., Rudy, Y. (2001). Models of cardiac ventricular action potentials: iterative interaction between experiment and simulation. Philosophical Transactions of the Royal Society of London. Series A: Mathematical, Physical and Engineering Sciences 359: 1127-1142. Nogami, A. (2011a). Purkinje-related arrhythmias part I: monomorphic ventricular tachycardias. Pacing Clin Electrophysiol 34: 624-650. Nogami, A. (2011b). Purkinje-related arrhythmias part ii: polymorphic ventricular tachycardia and ventricular fibrillation. Pacing Clin Electrophysiol 34: 1034-1049. Piazza, N., de Jaegere, P., Schultz, C., Becker, A. E., Serruys, P. W., Anderson, R. H. (2008). Anatomy of the aortic valvar complex and its implications for transcatheter implantation of the aortic valve. Circ Cardiovasc Interv 1: 74-81. Pick, A., Langendorf, R. (1974). The dual function of the A-V junction. Am Heart J 88: 790-797. Pop, M., Sermesant, M., Liu, G., Relan, J., Mansi, T., Soong, A., Peyrat, J. M., Truong, M. V., Fefer, P., McVeigh, E. R., Delingette, H., Dick, A. J., Ayache, N., Wright, G. A. (2012). Construction of 3D MR image-based computer models of pathologic hearts, augmented with histology and optical fluorescence imaging to characterize action potential propagation. Med Image Anal 16: 505-523. 122 Prystowsky, E. N. (1997). Atrioventricular node reentry: physiology and radiofrequency ablation. Pacing Clin Electrophysiol 20: 552-571. Rehak, R., Bartoletti, T. M., Engbers, J. D., Berecki, G., Turner, R. W., Zamponi, G. W. (2013). Low voltage activation of KCa1.1 current by Cav3-KCa1.1 complexes. PLoS One 8: e61844. Rolls, G. (2012). "The Process of Fixation and the Nature of Fixatives." Fixation and Fixatives Retrieved 20/06/2014, from http://www.leicabiosystems.com/pathologyleaders/fixation-and-fixatives-1-theprocess-of-fixation-and-the-nature-of-fixatives/. Sanchez-Quintana, D., Cabrera, J. A., Farre, J., Climent, V., Anderson, R. H., Ho, S. Y. (2005). Sinus node revisited in the era of electroanatomical mapping and catheter ablation. Heart 91: 189-194. Sawaya, F., Liff, D., Stewart, J., Lerakis, S., Babaliaros, V. (2012). Aortic stenosis: a contemporary review. Am J Med Sci 343: 490-496. Schwarz, F., Baumann, P., Manthey, J., Hoffmann, M., Schuler, G., Mehmel, H. C., Schmitz, W., Kubler, W. (1982). The effect of aortic valve replacement on survival. Circulation 66: 1105-1110. Seemann, G., Hoper, C., Sachse, F. B., Dossel, O., Holden, A. V., Zhang, H. (2006). Heterogeneous three-dimensional anatomical and electrophysiological model of human atria. Philos Transact A Math Phys Eng Sci 364: 1465-1481. Semmler, W., Schwaiger, M. (2008). Molecular Imaging I, Springer. Shi, W., Wymore, R., Yu, H., Wu, J., Wymore, R. T., Pan, Z., Robinson, R. B., Dixon, J. E., McKinnon, D., Cohen, I. S. (1999). Distribution and prevalence of hyperpolarization-activated cation channel (HCN) mRNA expression in cardiac tissues. Circ Res 85: e1-6. Silverman, M. E., Upshaw, C. B., Jr. (2002). Walter Gaskell and the understanding of atrioventricular conduction and block. J.Am.Coll.Cardiol. 39: 1574-1580. Sizarov, A., Devalla, H. D., Anderson, R. H., Passier, R., Christoffels, V. M., Moorman, A. F. (2011). Molecular analysis of patterning of conduction tissues in the developing human heart. Circ Arrhythm Electrophysiol 4: 532-542. Stannius, H. (1852). Versuche am Froschherzen, Veit. Stephenson, R. S., Boyett, M. R., Hart, G., Nikolaidou, T., Cai, X., Corno, A. F., Alphonso, N., Jeffery, N., Jarvis, J. C. (2012). Contrast enhanced micro-computed tomography resolves the 3-dimensional morphology of the cardiac conduction system in mammalian hearts. PLoS One 7: e35299. 123 Stewart, B. F., Siscovick, D., Lind, B. K., Gardin, J. M., Gottdiener, J. S., Smith, V. E., Kitzman, D. W., Otto, C. M. (1997). Clinical factors associated with calcific aortic valve disease. Cardiovascular Health Study. J Am Coll Cardiol 29: 630-634. Tawara, S. (2000). The Conduction System of the Mammalian Heart: An Anatomico-histological Study of the Atrioventricular Bundle and the Purkinje Fibers, Imperial College Press. Truex, R. C., Smythe, M. Q. (1967). Reconstruction of the human atrioventricular node. Anat.Rec. 158: 11-19. Truex, R. C., Smythe, M. Q., Taylor, M. J. (1967). Reconstruction of the human sinoatrial node. Anat.Rec. 159: 371-378. Turner, R. W., Zamponi, G. W. (2014). T-type channels buddy up. Pflugers Arch 466: 661-675. van Dam, P. M., Proniewska, K., Maugenest, A. M., van Mieghem, N. M., Maan, A. C., de Jaegere, P. P., Bruining, N. (2014). Electrocardiographic imaging-based recognition of possible induced bundle branch blocks during transcatheter aortic valve implantations. Europace 16: 750-757. Vandael, D. H., Marcantoni, A., Mahapatra, S., Caro, A., Ruth, P., Zuccotti, A., Knipper, M., Carbone, E. (2010). Ca(v)1.3 and BK channels for timing and regulating cell firing. Mol Neurobiol 42: 185-198. Vhlab.umn.edu. (2014, 19/06/2013). "3D Modeling - Atlas of Human Cardiac Anatomy." Retrieved 20 Jun 2014, 2014, from http://www.vhlab.umn.edu/atlas/index.shtml. Xu, W., Liu, Y., Wang, S., McDonald, T., Van Eyk, J. E., Sidor, A., O'Rourke, B. (2002). Cytoprotective role of Ca2+- activated K+ channels in the cardiac inner mitochondrial membrane. Science 298: 1029-1033. Yanni, J., Boyett, M. R., Anderson, R. H., Dobrzynski, H. (2009). The extent of the specialized atrioventricular ring tissues. Heart Rhythm 6: 672-680. 124 PUBLICATIONS Full papers and book chapters Atkinson A, Hao G, Logantha S, Fedorenko O, Yanni J, Gilbert S, Benson A, Buckley D, Anderson R, Boyett M, Dobrzynski H. (2013) Functional, anatomical and molecular investigation of the atrioventricular ring tissue in relation to the cardiac conduction system in the rat heart. JAHA 2(6) e000246 (full paper) Atkinson A, Dobrzynski H, Boyett M R, Yanni J. Use of RealTime StatMiner in cardiac gene expression studies. Integromics White Paper. Integromics SL. 2013. www.integromics.com/wp-content/uploads/2013/10/StatMiner-in-Cardiac-GeneExpression1.pdf (white paper) Yanni J, Maczewski M, Mackiewicz U, Siew S, Fedorenko O, Atkinson A, Price M, Beresewicz A, Anderson R, Boyett M, Dobrzynski H. (2013) Functional and structural remodelling of the atrioventricular node and the atrioventricular ring tissue in heart failure. Histol Histopathol. (full paper) Logantha SJ, Atkinson A, Boyett MR, Dobrzynski H. (2014) Molecular basis of arrhythmias associated with the cardiac conduction system. Chapter 2: Cardiac arrhythmias: From basic mechanism to state of the art management, 1st edition, edited by Tinitou IC et al. Springer, London. (book chapter) Dobrzynski H, Anderson RH, Atkinson A, Borbas Z, D'Souza A, Fraser JF, Inada S, Logantha SJ, Monfredi O, Morris GM, Moorman AF, Nikolaidou T, Schneider H, Szuts V, Temple IP, Yanni J, Boyett MR. (2013) Structure, function and clinical relevance of the cardiac conduction system, including the atrioventricular ring and outflow tract tissues. Pharmacol Ther. 139:260-88 (review article) Abstracts Heart Rhythm 2014. Atkinson A, Anderson RH, Boyett MR, Bateman MG, Iaizzo PA, Dobrzynski H. 3D anatomy of the human cardiac conduction system and its relationship to the aortic valve. AB22-03. 125 Heart Rhythm 2014. Logantha SJRJ, Stokke MK, Atkinson AJ, Parveen S, Sjaastad I, Sejersted OM, Boyett MR, Dobrzynski H. Sinoatrial node pacemaking is disrupted in the SERCA2 knockout mouse. PO03-01 IUPS 2013. Atkinson A, Nikolaidou T, Iaizzo PA, Molenaar P, Boyett MR, Dobrzynski H. 3D reconstruction of the human cardiac conduction system based on micro-CT and histology. IUPS 2013. Logantha S, Schneider H, Atkinson A, Hao G, Boyett M, Dobrzynski H. Pacemaker phenotype is the basis of tachyarrhythmias originating in the atrioventricular rings and the right ventricular outflow tract. IUPS 2013. Xiao Y, Cai X, Atkinson A, Logantha S, Hart G, Boyett M, Shui Z, Dobrzynski H. Histological and immunohistochemical study of the myocardial sleeves of the extensive pulmonary vein network of the rat. BCS 2013. Saeed Y, Borbas Z, Temple I, Atkinson A, Yanni J, Boyett M, Garratt C, Dobrzynski H. Ageing is associated with changes in structure and ion channel expression within the atrioventricular conduction axis. Heart 2013; 99: A139. 126 APPENDIX 127 Figure A1. Sample of outlined images used for sinus node reconstruction. Images from different levels of SN sample from inferior (1) to superior (12). Atrial muscle (red), paranodal area (green) and SN (yellow) outlined. 128 Figure A2. Comparison of whole heart histology and CT images. A. Masson’s trichrome histology section. B. CT image from equivalent level of the heart. 129 Figure A3. Computer simulation created using the whole heart 3D anatomical reconstruction (S. Kharche, unpublished data). 130 KCa1.1 KCa1.1 Vimentin α-actin 20 µm Figure A4. KCa1.1 Localisation. Double labelling of KCa1.1 (green) with Vimentin (left; red) and α-actin (right; red). 131