* Your assessment is very important for improving the work of artificial intelligence, which forms the content of this project
Download SOLUTE TRANSPORT
Survey
Document related concepts
Transcript
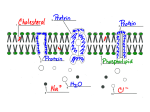
Bio 119 Solute Transport 7/11/2004 SOLUTE TRANSPORT READING: BOM-10 Sec. 4.7 Membrane Transport Systems p. 71 DISCUSSION QUESTIONS BOM-10: Chapter 4; #6, #8 1. What are the 4 essential features of carrier mediated transport? 2. What does it mean to say that "carrier mediated transport mechanisms exhibit saturation kinetics"? 3. What are two essential features of active transport? 4. What argument/s can be produced to rationalize the relative prevalence of passive transport mechanisms in higher eukaryotes compared to the prevalence of active transport mechanisms in bacteria? 6. In what two fundamentally different ways is energy coupled to active transport? 7. How does group translocation violate the strict definition of active transport? 8. How is the capacity to transport sugars maintained by bacteria even when they are energy limited? 10. Characterize each of the transport systems diagrammed below. [antiport vs. symport; electroneutral vs. electrogenic] OUT IN HSO4- H+ H+ lactose H+ Na+ 1 of 5 Bio 119 Solute Transport 7/11/2004 GENERAL In E. coli, the genome contains 427 genes for transport proteins, appox. 10% of the entire genome. Passive Diffusion vs. Protein Carrier Mediated Transport Solutes entering cell by passive diffusion. Solutes requiring carriers. Solute H 2O O2 N2 CO2 glycerol urea tryptone glucose ClNa+ Permeability Coefficient mol/sec cm2 10-2 10-6 10-6 10-7 10-7 10-10 10-12 ESSENTIAL FEATURES OF PROTEIN CARRIER MEDIATED TRANSPORT 1. High degree of solute specificity. 2. Competitive inhibition (by substrate analogs; may exert bacteriostatic effects) 3. Inducible by specific solutes (transcriptional regulation predominant) 4. Abolished by specific mutations in genes coding for transport proteins(some mutations may eliminate energy coupling, thus converting active transport to facilitated diffusion) 5. Saturation kinetics (carrier saturation is unusual in nature) Vmax Vmax/2 Km [So]-[Si] Facilitated Diffusion vs Active Transport For bacteria, intracellular solute concentrations are typically higher than extracellular concentrations. This is in contrast to the cells of multicellular eukaryotes, which are typically bathed in fluids whose 2 of 5 Bio 119 Solute Transport 7/11/2004 solute content is maintained at high levels by the organisms itself. Therefore, microorganisms depend more on active transport while higher eukaryotes frequently use passive diffusion. ESSENTIAL FEATURES OF ACTIVE TRANSPORT 1.[S] in > [S] out 2.Energy Coupling Formally, energy used to alter Km in < Km out, thereby shifting [Si] > [So] I. II. PASSIVE DIFFUSION Equilibrium State Energy Coupled ? [S] in = [S] out NO CARRIER MEDIATED TRANSPORT A. FACILITATED DIFFUSION [S] in = [S] out NO B. GROUP TRANSLOCATION [S*] in > [S] out YES/NO C. ACTIVE TRANSPORT [S] in > [S] out YES 1. Simple Transport 2. ABC System (ATP-Dependent) SIMPLE TRANSPORT 3 of 5 Bio 119 Solute Transport 7/11/2004 uniport vs. symport vs. antiport electroneutral vs. electrogenic typically 12 trans-membrane alpha-helices OUT + K HSO 4 lactose + HSO 4 lactose + Na PM IN K + Na + [H ] electroneutral antiport [MASS] electrogenic symport [ELECTRICAL + MASS] electroneutral symport [MASS] electrogenic uniport [ELECTRICAL] “ABC” ATP-DEPENDENT TRANSPORT typically involve periplasmic binding proteins examples: arg,his,leu gal,malt, rib Experimental distinction from ∆p-coupled active transport: blocked by ionophoric uncouplers rather than ATPase inhibitors GROUP TRANSLOCATION Group translocation systems are unique to bacteria. In a group translocation process, the solute is chemically altered (by phosphorylation, for example) as it is transported into the cell. The altered form of the solute is typically more ionic than original solute and therefore is likely to have lower membrane solubility. The carrier protein does not bind/transport the altered solute as efficiently as the original solute. Although the altered solute thus accumulates at high concentration in the cell relative to the exterior concentration of unaltered solute, this does not fit the strict definition of active transport. The altered solute is trapped in the cell and "cheats" the concentration gradient. Moreover, the modification of the solute is usually the first step in its biochemical utilization of the substrate. Sugar Phosphotransferase System of Gram-Negative Bacteria is classic example: 4 of 5 Bio 119 Solute Transport 7/11/2004 Note that this process is energy coupled in the sense that a high energy phosphate bond in PEP that could otherwise phosphorylate ADP is used instead to provide for the phosphorylation of the glucose substrate. Additionally, note that energy is conserved in the sense that the phosphorylation would be required (at the expense of an ATP) anyway by the first step of glucose catabolism. Other sugars (mannose, fructose, N-acetylglucosamine, -glucosides) are also transported by this system. E II proteins are specific for each sugar and are specifically induced, while HPr and E I are used for all substrates. E I and HPr mutants show facilitated diffusion for all substrates of the system. E Ia,b mutants show facilitated diffusion for a specific substrate. E IIc mutants fail to transport a specific substrate. During nutrient starvation [ATP] declines but glycolysis is regulated so that [PEP] remains relatively high. Thus when sugar substrates reappear, the system is poised to act. (Active transport systems would not be efficient under these conditions.) Other Group Translocation Systems in E. coli OUT IN free fatty acids HS - CoA fatty acyl - S - CoA purines/pyrimidines phosphoribosyl - PP nucleotide monophosphates + PP 5 of 5