Characterisation of the katA gene encoding a catalase and evidence
... The deduced amino acid sequence of the truncated ORF1, designated all, is similar to allantoinases that are implicated in the assimilation of allantoin (purine catabolism) [18]. ORF2 is located on the same strand as katA and shared no signi¢cant similarity with any sequence in the databases. The ded ...
... The deduced amino acid sequence of the truncated ORF1, designated all, is similar to allantoinases that are implicated in the assimilation of allantoin (purine catabolism) [18]. ORF2 is located on the same strand as katA and shared no signi¢cant similarity with any sequence in the databases. The ded ...
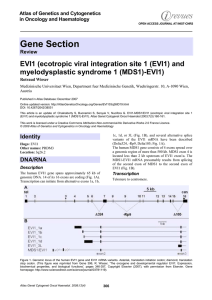
Gene Section EVI1 (ecotropic viral integration site 1 (EVI1) and
... EVI1 is thought to exert its biological functions mainly by acting as a transcription factor. In addition, however, EVI1 has been reported to inhibit c-jun Nterminal kinase, and to stimulate PI3K/AKT signalling. ...
... EVI1 is thought to exert its biological functions mainly by acting as a transcription factor. In addition, however, EVI1 has been reported to inhibit c-jun Nterminal kinase, and to stimulate PI3K/AKT signalling. ...
Characterization of chaperonin 10 (Cpn10)
... Identification, isolation and characterization of the E. histolytica gene encoding Cpn10 Searches of preliminary data generated by the E. histolytica genome project at the Sanger Institute revealed several clones with sequence similarity to the human Cpn10 protein sequence. PCR primers based on thes ...
... Identification, isolation and characterization of the E. histolytica gene encoding Cpn10 Searches of preliminary data generated by the E. histolytica genome project at the Sanger Institute revealed several clones with sequence similarity to the human Cpn10 protein sequence. PCR primers based on thes ...
Origin and Evolution of a New Gene Descended From alcohol
... mulleri, the putative Adh pseudogene is transcribed only in pupae and/or adults (FISCHERand MANIATIS 1985; SULLIVANet al. 1994), an expression pattern different from that of other Drosophila Adh genes (SULLIVAN et al. 1990). However, some aspects of the data are incompatible with a simple pseudogene ...
... mulleri, the putative Adh pseudogene is transcribed only in pupae and/or adults (FISCHERand MANIATIS 1985; SULLIVANet al. 1994), an expression pattern different from that of other Drosophila Adh genes (SULLIVAN et al. 1990). However, some aspects of the data are incompatible with a simple pseudogene ...
Practical: Ranges
... names(genes) <- c("FBgn0039155", "FBgn0085359") genes # now with names ## GRanges with 2 ranges and 0 metadata columns: ...
... names(genes) <- c("FBgn0039155", "FBgn0085359") genes # now with names ## GRanges with 2 ranges and 0 metadata columns: ...
Protein kinases of the human malaria parasite Plasmodium
... development in the mosquito involves a process of sporogony, producing sporozoites that accumulate in the salivary glands and are now ready to infect a new human host (see http://www.malaria.org for information on malaria). The observation that many parasitic ePKs display profound structural and fun ...
... development in the mosquito involves a process of sporogony, producing sporozoites that accumulate in the salivary glands and are now ready to infect a new human host (see http://www.malaria.org for information on malaria). The observation that many parasitic ePKs display profound structural and fun ...
Is HP1 an RNA detector that functions both in repression and
... may suggest roles for HP1 in regulating gene expression after the initiation of transcription. At 3 h after heat shock induction, transcript and protein levels were decreased in loss of function mutants but increased in gain of function mutants. The reverse was observed 7 h after heat shock. The aut ...
... may suggest roles for HP1 in regulating gene expression after the initiation of transcription. At 3 h after heat shock induction, transcript and protein levels were decreased in loss of function mutants but increased in gain of function mutants. The reverse was observed 7 h after heat shock. The aut ...
Rhizopus Raw-Starch-Degrading Glucoamylase: Its
... the yeast DNAmoiety. IR indicates the inverted repeat sequence of 2 /im DNA.The restriction sites P, H, B, and Pv indicates Pstl, Hindlll, BamHl and Pvull, respectively. ...
... the yeast DNAmoiety. IR indicates the inverted repeat sequence of 2 /im DNA.The restriction sites P, H, B, and Pv indicates Pstl, Hindlll, BamHl and Pvull, respectively. ...
Journal of Bacteriology
... a diminished and delayed but significant Ini phenotype. To investigate the roles of the nodF and nodE genes in the induction of Ini, nodF: :Tn5 (strain RBL5657) and nodE: :Tn5 (strain RBL5602) mutants were complemented with an IncP plasmid harboring either a cloned nodFE or a cloned nodE gene under ...
... a diminished and delayed but significant Ini phenotype. To investigate the roles of the nodF and nodE genes in the induction of Ini, nodF: :Tn5 (strain RBL5657) and nodE: :Tn5 (strain RBL5602) mutants were complemented with an IncP plasmid harboring either a cloned nodFE or a cloned nodE gene under ...
Unit F215 - Control, genomes and environment - Medium band
... production of an enzyme that will convert lactose into a compound that can enter the respiratory pathway. When glucose is present, this gene is not expressed but when it is not present the enzyme is made. This is because a gene isn’t just on its own. Each gene also has two lengths of DNA, known as t ...
... production of an enzyme that will convert lactose into a compound that can enter the respiratory pathway. When glucose is present, this gene is not expressed but when it is not present the enzyme is made. This is because a gene isn’t just on its own. Each gene also has two lengths of DNA, known as t ...
Optimizing cofactor availability for the production of recombinant
... from strains producing HRP alone was set to 100%. In addition to strong constitutive co-overexpression of either HEM1 or HEM3 from PGAP, we also tested co-overexpression of these two genes from either PAOX1 or PCAT1. Both promoters are strongly methanol-inducible, however PCAT1 is already active in ...
... from strains producing HRP alone was set to 100%. In addition to strong constitutive co-overexpression of either HEM1 or HEM3 from PGAP, we also tested co-overexpression of these two genes from either PAOX1 or PCAT1. Both promoters are strongly methanol-inducible, however PCAT1 is already active in ...
Characterization of Saccharomyces cerevisiae deficient in
... In the present study we have identified a gene required for the expression of PLD activity in yeast. Cells lacking expression of this gene show a complete lack of PLD activity in our in itro assay system. While it is possible that yeast expresses other forms of PLD that are not detected by this par ...
... In the present study we have identified a gene required for the expression of PLD activity in yeast. Cells lacking expression of this gene show a complete lack of PLD activity in our in itro assay system. While it is possible that yeast expresses other forms of PLD that are not detected by this par ...
Developmental Regulation of Genes Encoding Universal Stress
... Smp_136870 and Smp_136890) have been analyzed in the context of a phylogenomic analysis of metazoan USPs.18 Further, the functional annotations integrated into the relational database SchistoDB (http:// www.schistodb.net) include gene expression data documented on its developmental stages from hight ...
... Smp_136870 and Smp_136890) have been analyzed in the context of a phylogenomic analysis of metazoan USPs.18 Further, the functional annotations integrated into the relational database SchistoDB (http:// www.schistodb.net) include gene expression data documented on its developmental stages from hight ...
Detection and copy number estimation of the transgenic nucleotide
... these rice varieties can effectively reduce the loss and stabilize rice yield [3]. With the rapid improvements in genetic engineering, transgenic rice with a number of genes encoding different proteins, such as Bacillus thuringiensis (Bt), protease inhibitors [4], and plant lectins [5], has been dev ...
... these rice varieties can effectively reduce the loss and stabilize rice yield [3]. With the rapid improvements in genetic engineering, transgenic rice with a number of genes encoding different proteins, such as Bacillus thuringiensis (Bt), protease inhibitors [4], and plant lectins [5], has been dev ...
Interactions Between Genes Controlling Pathogenicity in the Flax
... strains CH 5 and I both being heterozygous for the postulated inhibitor gene and either heterozygous or homozygous dominant for the AM,/aM, gene pair. If both parents are IM'dMi AMIaM, a 3:13 (avirulent:virulent) ratio would be expected in all three families. If both are IM, iM, AM, AM, a 1:3 ratio ...
... strains CH 5 and I both being heterozygous for the postulated inhibitor gene and either heterozygous or homozygous dominant for the AM,/aM, gene pair. If both parents are IM'dMi AMIaM, a 3:13 (avirulent:virulent) ratio would be expected in all three families. If both are IM, iM, AM, AM, a 1:3 ratio ...
Analysis of DNA transcription termination sequences of gene coding
... major mechanisms (von Hippel 1998). One of them depends on Rho, which is a protein blocking RNA synthesis at specific sites. Rho, encoded by the gene rho, is a homohexamer, which has ATPase activity. The second one, called Rhoindependent termination (or intrinsic termination), is related with presen ...
... major mechanisms (von Hippel 1998). One of them depends on Rho, which is a protein blocking RNA synthesis at specific sites. Rho, encoded by the gene rho, is a homohexamer, which has ATPase activity. The second one, called Rhoindependent termination (or intrinsic termination), is related with presen ...
here - Genetics
... as clearing-houses for this purpose appear regularly in the Microbial Genetics Bulletin. C. Alleles. The nature of any particular mutational change is not indicated by the genotypic symbol. For example, araBl might be a base-pair substitution or a small deletion. In each case, however, the symbol in ...
... as clearing-houses for this purpose appear regularly in the Microbial Genetics Bulletin. C. Alleles. The nature of any particular mutational change is not indicated by the genotypic symbol. For example, araBl might be a base-pair substitution or a small deletion. In each case, however, the symbol in ...
In vivo evidence for the prokaryotic model of extended codon
... binding site or on nucleotides following the initiation codon, which may (Moll et al., 2001) constitute a regulatory `downstream box'. What has not been reassessed is the possibility of extended interactions between the mRNA and the initiator tRNA (tRNAfMet). The question of extended tRNAfMet±mRNA b ...
... binding site or on nucleotides following the initiation codon, which may (Moll et al., 2001) constitute a regulatory `downstream box'. What has not been reassessed is the possibility of extended interactions between the mRNA and the initiator tRNA (tRNAfMet). The question of extended tRNAfMet±mRNA b ...
Problem Sets - MIT Biology
... have isolated three mutations, “a,” “b” and “d,” each of which causes the same phenotype. When you mate a strain containing any one of these three mutations to wild-type, the resulting diploid exhibits the wild-type phenotype. You are in the process of doing complementation tests with these mutants. ...
... have isolated three mutations, “a,” “b” and “d,” each of which causes the same phenotype. When you mate a strain containing any one of these three mutations to wild-type, the resulting diploid exhibits the wild-type phenotype. You are in the process of doing complementation tests with these mutants. ...
Blankety Blank - misslongscience
... Blankety Blank 2. A gene is a sequence of nucleotides along a piece of DNA that determines a single characteristic of an organism. It does this by coding for particular polypeptides that make up the enzymes needed in a biochemical pathway. ...
... Blankety Blank 2. A gene is a sequence of nucleotides along a piece of DNA that determines a single characteristic of an organism. It does this by coding for particular polypeptides that make up the enzymes needed in a biochemical pathway. ...
::: Gene Set Enrichment Analysis - GSEA
... The Kolmogorov–Smirnov test is used to determine whether two underlying one-dimensional probability distributions differ, or whether an underlying probability distribution differs from a hypothesized distribution, in either case based on finite samples. The one-sample KS test compares the empirica ...
... The Kolmogorov–Smirnov test is used to determine whether two underlying one-dimensional probability distributions differ, or whether an underlying probability distribution differs from a hypothesized distribution, in either case based on finite samples. The one-sample KS test compares the empirica ...
ARF-Aux/IAA interactions through domain III/IV are not strictly
... on auxin response genes.9 Other results suggest that the reversal of this process (i.e., induction Figure 4. Dose response for 1-NAA on the expression of an integrated 2XD0:GUS auxinof an active chromatin state through histone responsive reporter gene in Arabidopsis nph4-1/arf7 protoplasts transfect ...
... on auxin response genes.9 Other results suggest that the reversal of this process (i.e., induction Figure 4. Dose response for 1-NAA on the expression of an integrated 2XD0:GUS auxinof an active chromatin state through histone responsive reporter gene in Arabidopsis nph4-1/arf7 protoplasts transfect ...
The genetic epidemiology of idiopathic scoliosis
... to environmental influences [2]. Genetic complexity in IS is further inferred from inconsistent inheritance [3–6], discordance among monozygotic twins [7–9], and highly variable results from genetic studies. The standard of care for scoliosis has not changed significantly in the past three decades, ...
... to environmental influences [2]. Genetic complexity in IS is further inferred from inconsistent inheritance [3–6], discordance among monozygotic twins [7–9], and highly variable results from genetic studies. The standard of care for scoliosis has not changed significantly in the past three decades, ...
Glioma heterogeneity and the LAT-1
... Glioma heterogeneity and the L-Amino acid transporter-1 (LAT1): A first step to stratified BPA-based BNCT? ...
... Glioma heterogeneity and the L-Amino acid transporter-1 (LAT1): A first step to stratified BPA-based BNCT? ...