Structural biology and drug design
... QSAR – Qantitative Structure-Activity Relationship • More advanced ligand-based drug design strategy/method • In addition to a set of ligands, biochemical activity data must be availible • Uses statistical methods (multi-variable regression) to relate a set of "predictor" variables (X) to the poten ...
... QSAR – Qantitative Structure-Activity Relationship • More advanced ligand-based drug design strategy/method • In addition to a set of ligands, biochemical activity data must be availible • Uses statistical methods (multi-variable regression) to relate a set of "predictor" variables (X) to the poten ...
Topics in Protein-Protein Docking
... • Docking two molecules means constructing the coordinates of the bound state. • Bound state is called the complex. • We require coordinates for the independent molecules as input • Molecules move towards each other and bind/‘dock’ • But aim is to predict their docked configuration (not describe th ...
... • Docking two molecules means constructing the coordinates of the bound state. • Bound state is called the complex. • We require coordinates for the independent molecules as input • Molecules move towards each other and bind/‘dock’ • But aim is to predict their docked configuration (not describe th ...
Chemistry B11 Chapter 4 Chemical reactions
... Chemical Equation: we represent a chemical reaction in the form of a chemical equation, using chemical formulas for the reactants and products, and an arrow to indicate the direction in which the reaction proceeds. Note: It is important to show the state of each reactant and product in a chemical eq ...
... Chemical Equation: we represent a chemical reaction in the form of a chemical equation, using chemical formulas for the reactants and products, and an arrow to indicate the direction in which the reaction proceeds. Note: It is important to show the state of each reactant and product in a chemical eq ...
Analyzing ITC Data for the Enthalpy of Binding Metal Ions to Ligands
... The enthalpy, ΔHITC, reported from the fit of titration curve represents the overall heat in the reaction cell. In order to account for all of the processes during a reaction it is recommended to analyze the data and account for each separate chemical event. Analysis may seem overwhelming at first, ...
... The enthalpy, ΔHITC, reported from the fit of titration curve represents the overall heat in the reaction cell. In order to account for all of the processes during a reaction it is recommended to analyze the data and account for each separate chemical event. Analysis may seem overwhelming at first, ...
Integration of Photosynthetic Protein Molecular Complexes in Solid
... *The photosynthetic complexes may be used as an interfacial material in photovoltaic devices. *Evolved within a thin membrane interface, photosynthetic complexes sustain large open circuit voltages of 1.1V without significant electron-hole recombination, and they may be self-assembled into an insula ...
... *The photosynthetic complexes may be used as an interfacial material in photovoltaic devices. *Evolved within a thin membrane interface, photosynthetic complexes sustain large open circuit voltages of 1.1V without significant electron-hole recombination, and they may be self-assembled into an insula ...
Chemiluminescent and Fluorescent Westerns
... in a broad spectra, emission wavelength cannot be used to differentiate signal from individual proteins. Thus, protein differentiation necessitates probing for proteins that are easily resolved during the electrophoresis process. This can become problematic when detecting co-migrating proteins, spec ...
... in a broad spectra, emission wavelength cannot be used to differentiate signal from individual proteins. Thus, protein differentiation necessitates probing for proteins that are easily resolved during the electrophoresis process. This can become problematic when detecting co-migrating proteins, spec ...
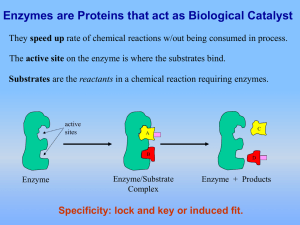
2. Enzymes
... 2) Co-factors: Inorganic components required for substrate binding at the active site. e.g. Ca2+, Mg2+ or Cu2+ (conformational changes). 3) Co-enzymes: Small organic molecules to accept and transfer electrons (e-s) from different enzymatic reactions. e.g. NAD+ shuttles e-s in glycolysis. ...
... 2) Co-factors: Inorganic components required for substrate binding at the active site. e.g. Ca2+, Mg2+ or Cu2+ (conformational changes). 3) Co-enzymes: Small organic molecules to accept and transfer electrons (e-s) from different enzymatic reactions. e.g. NAD+ shuttles e-s in glycolysis. ...
Enzymes: “Helper” Protein molecules
... Enzymes are not changed by the reaction used only temporarily re-used again for the same reaction with other molecules very little enzyme needed to help in many reactions ...
... Enzymes are not changed by the reaction used only temporarily re-used again for the same reaction with other molecules very little enzyme needed to help in many reactions ...
Chapter 6 "Mechanisms of Enzymes" Reading Assignment: pp. 158
... transition state has a structure between that of the reactant and the product. Note that the tetrahedral intermediate formed in Reaction 6.1 also is produced via a transition state structure (not shown). In this case, the structure of the transition state and the intermediate are thought to be very ...
... transition state has a structure between that of the reactant and the product. Note that the tetrahedral intermediate formed in Reaction 6.1 also is produced via a transition state structure (not shown). In this case, the structure of the transition state and the intermediate are thought to be very ...
Chapter 8 - Chemical Equations
... * because there is oxygen in every compound in the equation, it may be helpful to count the number of a polyatomic ion, rather than splitting the polyatomic ion into its elements and then counting.* EXAMPLE #6: ...
... * because there is oxygen in every compound in the equation, it may be helpful to count the number of a polyatomic ion, rather than splitting the polyatomic ion into its elements and then counting.* EXAMPLE #6: ...
Complete ionic equation
... • For each element, the number of atoms on the reactant side must equal the number of atoms on the product side. • The subscripts cannot change. Only coefficients can be changed. • The coefficients must be whole numbers. • The coefficients must be simplified (divided down) as much as possible. ...
... • For each element, the number of atoms on the reactant side must equal the number of atoms on the product side. • The subscripts cannot change. Only coefficients can be changed. • The coefficients must be whole numbers. • The coefficients must be simplified (divided down) as much as possible. ...
Chapter 17 Thermodynamics: Directionality of Chemical Reactions
... Most exothermic reactions are product favored. • E is transferred to the surroundings. • Bond (potential) E dispersed into many more atoms ...
... Most exothermic reactions are product favored. • E is transferred to the surroundings. • Bond (potential) E dispersed into many more atoms ...
1.4 enzymes 2014
... ALL MUST… Know that enzymes are biological catalysts that speed up the rate of chemical reactions and are made of ...
... ALL MUST… Know that enzymes are biological catalysts that speed up the rate of chemical reactions and are made of ...
Reaction Analysis and PAT Tools
... ReactIR collects data in the mid infrared spectral region, which provides characteristic fingerprint region bands that are associated with fundamental vibrations in the molecules of interest. iC IR takes this spectral data and converts it to reaction information that is then used for understanding t ...
... ReactIR collects data in the mid infrared spectral region, which provides characteristic fingerprint region bands that are associated with fundamental vibrations in the molecules of interest. iC IR takes this spectral data and converts it to reaction information that is then used for understanding t ...
Charge transport in a polypeptide chain
... transfer processes along pure polypeptides, without attaching donors or acceptors. They employed only natural amino acids where one of these amino acids contains a natural chromophore, i.e. trypotophan. Charge is introduced into the system via photoexcitation. In their experiment, they found that ch ...
... transfer processes along pure polypeptides, without attaching donors or acceptors. They employed only natural amino acids where one of these amino acids contains a natural chromophore, i.e. trypotophan. Charge is introduced into the system via photoexcitation. In their experiment, they found that ch ...
CHEM 30
... -calculate rate of reactions using: r = Δc/Δt - explain the factors which effect reaction rates; predicting rate of reactions - concentration vs time graphs – instantaneous rate of reaction - use activation energy diagrams and kinetic energy diagrams to show effect of temperature and catalysts on re ...
... -calculate rate of reactions using: r = Δc/Δt - explain the factors which effect reaction rates; predicting rate of reactions - concentration vs time graphs – instantaneous rate of reaction - use activation energy diagrams and kinetic energy diagrams to show effect of temperature and catalysts on re ...
spring semester review
... (a) Shifts right (b) shifts left (c) no change (d) can’t tell from the information given 6. Add more Ni? 7. Add more CO (g) 8. Add Neon (g) 9. Remove Ni(CO)4 (g) 10. Cut volume by ½ 11. Increase Temperature 12. Add a catalyst 13. For which one of the following reactions is Kc equal to Kp? (a) H2(g) ...
... (a) Shifts right (b) shifts left (c) no change (d) can’t tell from the information given 6. Add more Ni? 7. Add more CO (g) 8. Add Neon (g) 9. Remove Ni(CO)4 (g) 10. Cut volume by ½ 11. Increase Temperature 12. Add a catalyst 13. For which one of the following reactions is Kc equal to Kp? (a) H2(g) ...
1 Lecture 27: Metabolic Pathways Part I: Glycolysis
... Glyceraldehyde-3-P dehydrogenase: G-3-P + NAD+ → 1,3 bisphosphoglycerate + NADH This reaction proceeds in two steps. The first step is the oxidation of the aldehyde to the carboxylic acid (thioester) using NAD+ as the electron acceptor. This results in the formation of an covalent enzyme intermediat ...
... Glyceraldehyde-3-P dehydrogenase: G-3-P + NAD+ → 1,3 bisphosphoglycerate + NADH This reaction proceeds in two steps. The first step is the oxidation of the aldehyde to the carboxylic acid (thioester) using NAD+ as the electron acceptor. This results in the formation of an covalent enzyme intermediat ...
Equilibrium and Kinetic Studies of Ligand
... buffer or bis-tris (bis-[2-hydroxyethyl]iminotris [hydroxy ethyl]methane) buffer are used (9). The chromatograms of other metal-HQS complexes exhibited single peaks even when the eluents contained a phosphate buffer. We have shown that this anomalous behavior of the alu minum complex can be explain ...
... buffer or bis-tris (bis-[2-hydroxyethyl]iminotris [hydroxy ethyl]methane) buffer are used (9). The chromatograms of other metal-HQS complexes exhibited single peaks even when the eluents contained a phosphate buffer. We have shown that this anomalous behavior of the alu minum complex can be explain ...
22. Think of two different proteins: both are enzymes. a) What
... and what part of #39 does it relate to? When the pH level of the medium goes down when oxygen is present, it shows that mitochondria release protons. This supports the chemiosmotic mechanism (the link between electron transport and ATP synthesis= proton gradient). It helps to demonstrate the movemen ...
... and what part of #39 does it relate to? When the pH level of the medium goes down when oxygen is present, it shows that mitochondria release protons. This supports the chemiosmotic mechanism (the link between electron transport and ATP synthesis= proton gradient). It helps to demonstrate the movemen ...
06 Classification and modern methods of diagnostics
... The final product is available in high conc. and can be used for cloning (gene that has a product), diagnosis (gene of a virus) or fingerprinting (forensic investigation in crime scenes). ...
... The final product is available in high conc. and can be used for cloning (gene that has a product), diagnosis (gene of a virus) or fingerprinting (forensic investigation in crime scenes). ...
Slide 1
... molecules and join them together to form complex molecules. For example, glucose molecules go through a condensation reaction forming many glycosidic bonds between molecules and eventually forming the ...
... molecules and join them together to form complex molecules. For example, glucose molecules go through a condensation reaction forming many glycosidic bonds between molecules and eventually forming the ...