Cartoon modeling of proteins
... Ordered soup of ~1,000,000 different types of macromolecules Complex and specific network of interactions Ion channels and complexes the tip of the iceberg (croutons?) Much work on gene networks / intracellular pathways Mostly ignores spatial effects (well mixed pool / kinetics) ...
... Ordered soup of ~1,000,000 different types of macromolecules Complex and specific network of interactions Ion channels and complexes the tip of the iceberg (croutons?) Much work on gene networks / intracellular pathways Mostly ignores spatial effects (well mixed pool / kinetics) ...
Cartoon modeling of proteins
... Ordered soup of ~1,000,000 different types of macromolecules Complex and specific network of interactions Ion channels and complexes the tip of the iceberg (croutons?) Much work on gene networks / intracellular pathways Mostly ignores spatial effects (well mixed pool / kinetics) ...
... Ordered soup of ~1,000,000 different types of macromolecules Complex and specific network of interactions Ion channels and complexes the tip of the iceberg (croutons?) Much work on gene networks / intracellular pathways Mostly ignores spatial effects (well mixed pool / kinetics) ...
Why do simulations? - Course Website Directory
... the problem. The semi-empirical approach. But one cannot reliably extrapolate the model away from the empirical data. B. Maxwell, Boltzmann and Schrödinger gave us the model. All we must do is numerically solve the mathematical problem and determine the properties. (first-principles or ab initio met ...
... the problem. The semi-empirical approach. But one cannot reliably extrapolate the model away from the empirical data. B. Maxwell, Boltzmann and Schrödinger gave us the model. All we must do is numerically solve the mathematical problem and determine the properties. (first-principles or ab initio met ...
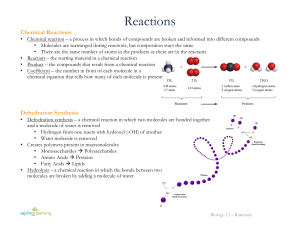
Reactions
... • Catalyst – a substance that affects the rate of a chemical reaction • Enzyme – a specialized protein that catalyzes biological reactions • Usually end with the suffix “-ase” and its name comes from the substrate • Have optimal temperatures and pH to maintain normal functioning • Enzymes lower the ...
... • Catalyst – a substance that affects the rate of a chemical reaction • Enzyme – a specialized protein that catalyzes biological reactions • Usually end with the suffix “-ase” and its name comes from the substrate • Have optimal temperatures and pH to maintain normal functioning • Enzymes lower the ...
Effect of ZnO on Pd/ZnO Catalysts in Steam Reforming of Methanol
... • L-proline has two active sites (an amino group and a ...
... • L-proline has two active sites (an amino group and a ...
The basis of specific ligand recognition by proteins
... fluctuating networks of interactions; these often involve water molecules with very short residence times. The general problem of mobility will be addressed by specifically looking at interfaces between proteins and DNA, where the protein binds specifically to a certain DNA sequence (“reading” of DN ...
... fluctuating networks of interactions; these often involve water molecules with very short residence times. The general problem of mobility will be addressed by specifically looking at interfaces between proteins and DNA, where the protein binds specifically to a certain DNA sequence (“reading” of DN ...
Headline Text 28 Point Color Text 2
... Chemical informatics and medicinal chemistry databases How molecular motors work Computer modeling in support of chemical and drug design • Polymer delivery systems • Catalysts • Small molecule drugs ...
... Chemical informatics and medicinal chemistry databases How molecular motors work Computer modeling in support of chemical and drug design • Polymer delivery systems • Catalysts • Small molecule drugs ...
Chemical Synthesis (sat6)
... A1: MgO and H2 -> Mg and H2O; A2: C and O2 -> CO2; A3: CO2 and H2O -> H2CO3; A4: MgO and H2 and O2 and C; minimize obj: H2CO3; Write(’Yes, H2CO3 is produced’); Write(’No, H2CO3 is not produced’); ...
... A1: MgO and H2 -> Mg and H2O; A2: C and O2 -> CO2; A3: CO2 and H2O -> H2CO3; A4: MgO and H2 and O2 and C; minimize obj: H2CO3; Write(’Yes, H2CO3 is produced’); Write(’No, H2CO3 is not produced’); ...
Lectures on Computational Biology
... Lectures on Computational Biology EFSS II National DongHua University Hualien, 2002 July 1-5 HC Lee Computational Biology Lab Center for Complex Systems & Biophysics National Central University ...
... Lectures on Computational Biology EFSS II National DongHua University Hualien, 2002 July 1-5 HC Lee Computational Biology Lab Center for Complex Systems & Biophysics National Central University ...
Syllabus
... There will be 12 homework assignments, a midterm and a final. All are given on a take-home basis. If you have any question about a problem before starting to work, you are strongly encouraged to discuss the matter with the professor or fellow students. That is, students are encouraged to discuss and ...
... There will be 12 homework assignments, a midterm and a final. All are given on a take-home basis. If you have any question about a problem before starting to work, you are strongly encouraged to discuss the matter with the professor or fellow students. That is, students are encouraged to discuss and ...
Fakulti: FAKULTI KEJURUTERAAN ELEKTRIK
... You must know theoretically the fabrication processes of a typical semiconductor device. In order to be familiarized with the TCAD simulation tool, refer to the manuals which are available in the lab. Silvaco's ATHENA is used to simulate the processing conditions for the formation of the device. Thi ...
... You must know theoretically the fabrication processes of a typical semiconductor device. In order to be familiarized with the TCAD simulation tool, refer to the manuals which are available in the lab. Silvaco's ATHENA is used to simulate the processing conditions for the formation of the device. Thi ...
how to deal accurately with both the core and valence electrons
... “A simple model can shed more light on Nature’s workings than a series of “ab-initio” calculations of individual cases, which,even if correct, are so-detailed that they hide reality instead of revealing it… A perfect computation simply reproduces Nature, it does not explain it.” ...
... “A simple model can shed more light on Nature’s workings than a series of “ab-initio” calculations of individual cases, which,even if correct, are so-detailed that they hide reality instead of revealing it… A perfect computation simply reproduces Nature, it does not explain it.” ...
Computational Modelling in Systems and Synthetic Biology
... modular patterns exhibiting robustness, signal filtering, amplification, adaption, error correction, etc. ...
... modular patterns exhibiting robustness, signal filtering, amplification, adaption, error correction, etc. ...
SYNTHESIS REACTIONS
... Before we go on to some examples, let's use this reaction to take care of some basic reaction terminology. The compound(s) on the left are called reactant(s). Those on the left are product(s). We separate different sets of reactants and products with + signs. We generally only use a single arrow ( → ...
... Before we go on to some examples, let's use this reaction to take care of some basic reaction terminology. The compound(s) on the left are called reactant(s). Those on the left are product(s). We separate different sets of reactants and products with + signs. We generally only use a single arrow ( → ...
Protein Folding and Modeling
... interacting via a realistic set of potentials based on the Lennard Jones potential and electrostatic Coulomb potential Includes correct bond lengths, bond angles, planar trans peptide bond, leads to faithful representation of protein geometry. ...
... interacting via a realistic set of potentials based on the Lennard Jones potential and electrostatic Coulomb potential Includes correct bond lengths, bond angles, planar trans peptide bond, leads to faithful representation of protein geometry. ...
Study Guide 2—Chemical Principles 1. Understand, define and be
... amino acid, peptide bond, primary structure, secondary structure, tertiary structure, quaternary structure, denature, nucleic acid, DNA, RNA, ATP, ADP. 2. Calculate the molecular weight of a given molecule, and describe how you would make a 1 molar solution using that molecule. 3. Describe how chemi ...
... amino acid, peptide bond, primary structure, secondary structure, tertiary structure, quaternary structure, denature, nucleic acid, DNA, RNA, ATP, ADP. 2. Calculate the molecular weight of a given molecule, and describe how you would make a 1 molar solution using that molecule. 3. Describe how chemi ...
Simulation of Prokaryotic Genetic Circuits
... Stochastic processes in regulatory kinetics Modelling macromolecular complexes Uncertainty in intracellular environment and reaction rates ...
... Stochastic processes in regulatory kinetics Modelling macromolecular complexes Uncertainty in intracellular environment and reaction rates ...
Modeling of Solid State Transformer for the FREEDM System
... The Solid State Transformer (SST) is an essential component in the FREEDM system and it would have made much more impact in the future smart grid. This report focuses on the modeling of SST and the controller hardware in the loop implementation of SST for the support of the FREEDM system demonstrati ...
... The Solid State Transformer (SST) is an essential component in the FREEDM system and it would have made much more impact in the future smart grid. This report focuses on the modeling of SST and the controller hardware in the loop implementation of SST for the support of the FREEDM system demonstrati ...
First Pass Monte Carlo Simulation of Basic Eukaryotic Cell Cycle
... potential for new incites into the dynamics of the control networks on a molecular level. Modeling with ODEs necessarily glosses over these dynamics because the basic assumption underlying the fitting of an ODE to a system implies that the reactant molecules exist in a well-mixed state for the durat ...
... potential for new incites into the dynamics of the control networks on a molecular level. Modeling with ODEs necessarily glosses over these dynamics because the basic assumption underlying the fitting of an ODE to a system implies that the reactant molecules exist in a well-mixed state for the durat ...
IT010 606Lxx Simulation and Modelling
... IT010 606 L01 SIMULATION AND MODELLING Teaching scheme ...
... IT010 606 L01 SIMULATION AND MODELLING Teaching scheme ...
Abstract in Microsoft Word® format 21318 bytes
... molecular scale. An important process for realizing this technology is the design of molecules for specific functions, such as wires, diodes and transistors. In this research area, the design of molecules with desirable properties for molecular devices is still a great challenge, since the behavior ...
... molecular scale. An important process for realizing this technology is the design of molecules for specific functions, such as wires, diodes and transistors. In this research area, the design of molecules with desirable properties for molecular devices is still a great challenge, since the behavior ...
Graduate Biochemistry 7.51: The Major Concepts
... As you will see from the syllabus, the lectures in this course are drawn from a wide range of topics in biochemistry. However, nearly all of the science we discuss is based on a discrete number of fundamental concepts that are common to most biochemical approaches. A major goal of this course is to ...
... As you will see from the syllabus, the lectures in this course are drawn from a wide range of topics in biochemistry. However, nearly all of the science we discuss is based on a discrete number of fundamental concepts that are common to most biochemical approaches. A major goal of this course is to ...
Project Details PPT
... oP R E V E N T W A S T E B Y U S I N G A SOLVENTLESS PROCESS. oM I N I M I Z E A M O U N T S O F S O L V E N T S A N D REAGENTS USED. ...
... oP R E V E N T W A S T E B Y U S I N G A SOLVENTLESS PROCESS. oM I N I M I Z E A M O U N T S O F S O L V E N T S A N D REAGENTS USED. ...