Assignment 3
... in the Water Chemistry text are taken into consideration. If a complex is in the Minteq.v4 database, you can use the log K that is in the database, even if it differs from the value in the Table. However, if a complex is absent from the database altogether, add it to the complexes that are considere ...
... in the Water Chemistry text are taken into consideration. If a complex is in the Minteq.v4 database, you can use the log K that is in the database, even if it differs from the value in the Table. However, if a complex is absent from the database altogether, add it to the complexes that are considere ...
Estimating the divisibility complex biological networks by
... Keywords: Biological networks, partitioning, graph sparseness, parallel computing Key points: 1. Biological networks represent the complexity of interactions among the components of natural systems (e.g. interactions among molecules in the cell); 2. Dividing biological networks into maximally discon ...
... Keywords: Biological networks, partitioning, graph sparseness, parallel computing Key points: 1. Biological networks represent the complexity of interactions among the components of natural systems (e.g. interactions among molecules in the cell); 2. Dividing biological networks into maximally discon ...
Toward detection of DNA-bound proteins using solid-state
... Toward detection of DNA-bound proteins using solid-state nanopores: insights from computer simulations Jeffrey Comer, Anthony Ho and Aleksei Aksimentiev* Department of Physics and Beckman Institute University of Illinois at Urbana-Champaign Urbana, Illinois, USA ...
... Toward detection of DNA-bound proteins using solid-state nanopores: insights from computer simulations Jeffrey Comer, Anthony Ho and Aleksei Aksimentiev* Department of Physics and Beckman Institute University of Illinois at Urbana-Champaign Urbana, Illinois, USA ...
CHE 4310 Fall 2011
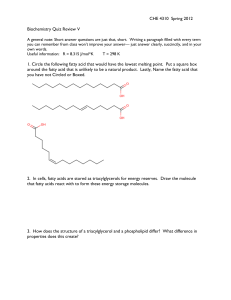
... 16. Describe the part of the glycolytic pathway from fructose 6-phosphate to glyceraldehyde 3phosphate. Show structures of intermediates, enzyme names, and indicate where any cofactors ...
... 16. Describe the part of the glycolytic pathway from fructose 6-phosphate to glyceraldehyde 3phosphate. Show structures of intermediates, enzyme names, and indicate where any cofactors ...
2.1 Molecules and metabolism
... • Molecular biology explains living processes in terms of the chemical substances involved. • Carbon atoms can form four covalent bonds allowing a diversity of stable compounds to exist. • Life is based on carbon compounds including carbohydrates, lipids, proteins and nucleic acids. • Metabolism is ...
... • Molecular biology explains living processes in terms of the chemical substances involved. • Carbon atoms can form four covalent bonds allowing a diversity of stable compounds to exist. • Life is based on carbon compounds including carbohydrates, lipids, proteins and nucleic acids. • Metabolism is ...
Biochemical Reactions
... Electrons are highly reactive and don’t exist on their own in cells If ...
... Electrons are highly reactive and don’t exist on their own in cells If ...
Introduction to ODE Modeling
... the gradient (gX,gY) of the chemoattractant molecules. P( x-> x+1, y->y) = ¼ + gX P( x-> x-1, y->y) = ¼ - gX P( x-> x, y->y+1) = ¼ + gY P( x-> x, y->y-1) = ¼ - gY Simulation similar to previous random walk.. ...
... the gradient (gX,gY) of the chemoattractant molecules. P( x-> x+1, y->y) = ¼ + gX P( x-> x-1, y->y) = ¼ - gX P( x-> x, y->y+1) = ¼ + gY P( x-> x, y->y-1) = ¼ - gY Simulation similar to previous random walk.. ...
Levels of organization in a living organism.
... What is Biochemistry? • A scientific discipline • Explains life at the molecular level • Explains biological forms and functions in chemical terms • Forms basis for studying other fields like cell biology, genetics, medicine, nutrition, immunology etc. • Closely relates to other discipline like che ...
... What is Biochemistry? • A scientific discipline • Explains life at the molecular level • Explains biological forms and functions in chemical terms • Forms basis for studying other fields like cell biology, genetics, medicine, nutrition, immunology etc. • Closely relates to other discipline like che ...
Designer enzymes Donald Hilvert ETH Zurich, Zurich, Switzerland
... structure-function relationships in these macromolecules is at best incomplete. Nature has solved the problem of protein design through the mechanism of Darwinian evolution. From primitive precursors, recursive cycles of mutation, selection and amplification of molecules with favorable traits have g ...
... structure-function relationships in these macromolecules is at best incomplete. Nature has solved the problem of protein design through the mechanism of Darwinian evolution. From primitive precursors, recursive cycles of mutation, selection and amplification of molecules with favorable traits have g ...
P systems–based Modelling of Cellular Signalling Pathways
... reasonable in some circumstances but not in many cases due to internal structure and low numbers and non–uniform distributions of certain key molecules in the cell. While differential equations models may produce useful results under certain conditions, they provide a rather incomplete view of what ...
... reasonable in some circumstances but not in many cases due to internal structure and low numbers and non–uniform distributions of certain key molecules in the cell. While differential equations models may produce useful results under certain conditions, they provide a rather incomplete view of what ...
Bio
... models of curriculum for educational programs. He has worked on the development of many simulation programs, most notably that of Serious Safety Event (SSE) training and facilitation courses for those learning how to debrief simulations. While Mike served as the Pediatric Advanced Life Support (PALS ...
... models of curriculum for educational programs. He has worked on the development of many simulation programs, most notably that of Serious Safety Event (SSE) training and facilitation courses for those learning how to debrief simulations. While Mike served as the Pediatric Advanced Life Support (PALS ...
BIOCHEMISTRY STUDY GUIDE Look over Chapter 3 Review on
... What is a functional group and what does it do to a compounds structure and function? How does the structure of phospholipids, linear molecules with a polar end and a nonpolar end, relate to their function in the cell membrane? Know what the monomers of each molecule of life are and examples of each ...
... What is a functional group and what does it do to a compounds structure and function? How does the structure of phospholipids, linear molecules with a polar end and a nonpolar end, relate to their function in the cell membrane? Know what the monomers of each molecule of life are and examples of each ...
Using Models to Represent Chemical Reactions
... filling models. See if you can identify how many of the models are molecular compounds, and how many are molecular elements. How could you tell? How many of the molecules are polar? How many are nonpolar? Which do you think has the highest boiling point? Why? Also, imagine that 4 molecules of methan ...
... filling models. See if you can identify how many of the models are molecular compounds, and how many are molecular elements. How could you tell? How many of the molecules are polar? How many are nonpolar? Which do you think has the highest boiling point? Why? Also, imagine that 4 molecules of methan ...
pathway_cell_models
... – data mining e.g. genome expression at gene or protein level contribute to conceptualisations of pathways ...
... – data mining e.g. genome expression at gene or protein level contribute to conceptualisations of pathways ...
fac Lecture Flyer-Anderson 4-20 - Chemical Engineering : University
... Two dimensional melting is a fascinating problem that demands powerful simulation tools and compute resources. Hexatic phases with quasi long range bond order and short range positional order require simulations of at least 1 million particles, which take 100's of hours to run on 64 GPUs of the Tita ...
... Two dimensional melting is a fascinating problem that demands powerful simulation tools and compute resources. Hexatic phases with quasi long range bond order and short range positional order require simulations of at least 1 million particles, which take 100's of hours to run on 64 GPUs of the Tita ...
Chapter 2 SWBATS Content Standards Cell Biology 1. The
... reactions that occur in specialized areas of the organism's cells. As a basis for understanding this concept: b. Students know enzymes are proteins that catalyze biochemical reactions without altering the reaction equilibrium and the activities of enzymes depend on the temperature, ionic conditions, ...
... reactions that occur in specialized areas of the organism's cells. As a basis for understanding this concept: b. Students know enzymes are proteins that catalyze biochemical reactions without altering the reaction equilibrium and the activities of enzymes depend on the temperature, ionic conditions, ...
The Chemicals of Life
... Base element for many macromolecules in the body Carbon is a small element that can form 4 stable covalent bonds with other atoms (look at valence electrons). The number of covalent bonds an atom can form is called its bonding capacity. Carbon can bond to other carbon atoms in straight chains, branc ...
... Base element for many macromolecules in the body Carbon is a small element that can form 4 stable covalent bonds with other atoms (look at valence electrons). The number of covalent bonds an atom can form is called its bonding capacity. Carbon can bond to other carbon atoms in straight chains, branc ...
Document
... Assuming that the number of relevant unfolded structures is proportional to the folding time, only 109 protein structures need to be simulated instead of 1090 structures. ...
... Assuming that the number of relevant unfolded structures is proportional to the folding time, only 109 protein structures need to be simulated instead of 1090 structures. ...
SysBio Class - The Computable Plant
... Software architecture to integrate data sources database model generator simulator optimizer hypothesis formulation e.g. SBML, Cellerator, KEGG, … Issue: How to span the gap between small- and genomic-scale data? Database (of reactions and pathway modeling information) Knowledge representati ...
... Software architecture to integrate data sources database model generator simulator optimizer hypothesis formulation e.g. SBML, Cellerator, KEGG, … Issue: How to span the gap between small- and genomic-scale data? Database (of reactions and pathway modeling information) Knowledge representati ...
A.P. Chemistry Complexation Reactions
... Fire It Up - Combustion Reactions – YouTube Double Replacement with Precipitates YouTube ...
... Fire It Up - Combustion Reactions – YouTube Double Replacement with Precipitates YouTube ...
Enzymes - Science Geek
... CA Standard Students know enzymes are proteins that catalyze biochemical reactions without altering the reaction equilibrium and the activities of enzymes depend on the temperature, ionic conditions, and the pH of the surroundings. ...
... CA Standard Students know enzymes are proteins that catalyze biochemical reactions without altering the reaction equilibrium and the activities of enzymes depend on the temperature, ionic conditions, and the pH of the surroundings. ...
Single molecule derivation of base pair free energies in
... allow for the controlled manipulation of individual molecules by exerting and detecting forces in the piconewton range1 . Molecular unzipping is a force-induced reaction that makes possible to disrupt the bonds that hold molecular structures in nucleic acids and proteins. In this way, for example, a ...
... allow for the controlled manipulation of individual molecules by exerting and detecting forces in the piconewton range1 . Molecular unzipping is a force-induced reaction that makes possible to disrupt the bonds that hold molecular structures in nucleic acids and proteins. In this way, for example, a ...