Efficient Sampling Methods for Protein Structure Refinement
... generate a structure prediction that improves upon a given homology model, especially in regions where a good template is not available. The conformational space of a protein is too large for an exhaustive search to be computationally feasible. An efficient sampling algorithm is therefore crucial to ...
... generate a structure prediction that improves upon a given homology model, especially in regions where a good template is not available. The conformational space of a protein is too large for an exhaustive search to be computationally feasible. An efficient sampling algorithm is therefore crucial to ...
Enzyme Notes Outline
... Proteins that speed up reactions without being changed themselves When are they used?: To speed up chemical reactions are too slow or have high activation energies ** Enzymes are very specific- they only catalyze one type of reaction ** What is the “substrate”-The reactants that interact with enzyme ...
... Proteins that speed up reactions without being changed themselves When are they used?: To speed up chemical reactions are too slow or have high activation energies ** Enzymes are very specific- they only catalyze one type of reaction ** What is the “substrate”-The reactants that interact with enzyme ...
republique française - Laboratoire Léon Brillouin (LLB)
... investigate solution organization or solvation dynamics give highly averaged information over all different kinds of molecular interactions, with a variety of time scales and length scales. To address these limitations, we have developed a well-defined model system based on amino acid monomers in so ...
... investigate solution organization or solvation dynamics give highly averaged information over all different kinds of molecular interactions, with a variety of time scales and length scales. To address these limitations, we have developed a well-defined model system based on amino acid monomers in so ...
Sharing Models
... representing cellular networks. A large number (>200) of tools support SBML. CellML: Stores models in mathematical form, therefore is quite general, but biological information is lost. Not possible to reconstruct network. Less than a hand-full of tools support CellML SBGN: A proposed standard for vi ...
... representing cellular networks. A large number (>200) of tools support SBML. CellML: Stores models in mathematical form, therefore is quite general, but biological information is lost. Not possible to reconstruct network. Less than a hand-full of tools support CellML SBGN: A proposed standard for vi ...
REPORT FOR EXPERIMENT 10 Double Displacement Reactions
... 1. Record your observations (Evidence of Reaction) of each experiment. Use the following terminology: (a) "Precipitate formed" (include the color), (b) "Gas evolved," (c) "Heat evolved," or (d) "No reaction observed." 2. Complete and balance the equation for each case in which a reaction occurred. F ...
... 1. Record your observations (Evidence of Reaction) of each experiment. Use the following terminology: (a) "Precipitate formed" (include the color), (b) "Gas evolved," (c) "Heat evolved," or (d) "No reaction observed." 2. Complete and balance the equation for each case in which a reaction occurred. F ...
Document
... A critical step in peptide synthesis is the activation of the carboxyl group for nucleophilic attack by an amine and consequent formation of a peptide bond. This activation can be achieved in a variety of ways - see examples on pg 44. In the example of peptide synthesis presented in class the carbox ...
... A critical step in peptide synthesis is the activation of the carboxyl group for nucleophilic attack by an amine and consequent formation of a peptide bond. This activation can be achieved in a variety of ways - see examples on pg 44. In the example of peptide synthesis presented in class the carbox ...
Intro to Biochemistry Pratt & Cornely Chapter 1
... compound is cold to the touch. What conclusions can you draw about the sign of the enthalpy change and the entropy change for this process? ...
... compound is cold to the touch. What conclusions can you draw about the sign of the enthalpy change and the entropy change for this process? ...
Unit 2 Test Review
... Organic compound that is the building block of organisms; made of amino acids Number (from 0-14) measuring the amount of hydrogen ions in a solution Molecules made during chemical reactions; on the right side of the equation A chemical that releases hydrogen ions in a solution Chemicals centered aro ...
... Organic compound that is the building block of organisms; made of amino acids Number (from 0-14) measuring the amount of hydrogen ions in a solution Molecules made during chemical reactions; on the right side of the equation A chemical that releases hydrogen ions in a solution Chemicals centered aro ...
Slide 1
... • A technique used to identify TFBS within a noncoding region of DNA of interest by comparing it to the orthologous sequences in different species (1988 by Tagle) • The function and DNA binding preferences of transcription factors are well-conserved between diverse species • Important non-coding DNA ...
... • A technique used to identify TFBS within a noncoding region of DNA of interest by comparing it to the orthologous sequences in different species (1988 by Tagle) • The function and DNA binding preferences of transcription factors are well-conserved between diverse species • Important non-coding DNA ...
Motor-clutch model for substrate stiffness sensing by living cells
... assess which model parameters control the stiffness at which sensing is optimal. Our exploration of parameter space reveals that no single parameter in the motor-clutch model can strongly control the setpoint for optimal stiffness sensing. Rather, parameters need to be changed coordinately to effect ...
... assess which model parameters control the stiffness at which sensing is optimal. Our exploration of parameter space reveals that no single parameter in the motor-clutch model can strongly control the setpoint for optimal stiffness sensing. Rather, parameters need to be changed coordinately to effect ...
Gizmos: Types of Reactions
... Question: How are chemical reactions classified? 1. Match: Most chemical reactions can be classified as one of four types. Using the chemical equations in the Gizmo as a guide, match the following definitions to the type of reaction. ____ One reactant is broken down into two or more products. ...
... Question: How are chemical reactions classified? 1. Match: Most chemical reactions can be classified as one of four types. Using the chemical equations in the Gizmo as a guide, match the following definitions to the type of reaction. ____ One reactant is broken down into two or more products. ...
Study Guide
... Carbon can form covalent bonds to 4 other atoms (simultaneously) Carbon can also form bonds to other carbons Any molecule that contains carbon is called an “organic molecule” o All organic molecules are combinations of: carbon, oxygen, hydrogen, phosphorus, and nitrogen B. Macromolecule- large ...
... Carbon can form covalent bonds to 4 other atoms (simultaneously) Carbon can also form bonds to other carbons Any molecule that contains carbon is called an “organic molecule” o All organic molecules are combinations of: carbon, oxygen, hydrogen, phosphorus, and nitrogen B. Macromolecule- large ...
Dicke_Profile_20100827
... fundamental understanding of electron transfer processes in organic molecules. Typically, we use Donor-Bridge-Acceptor (DBA) systems as model complexes to study the effect of different bridge structures on the charge transfer mechanism. ...
... fundamental understanding of electron transfer processes in organic molecules. Typically, we use Donor-Bridge-Acceptor (DBA) systems as model complexes to study the effect of different bridge structures on the charge transfer mechanism. ...
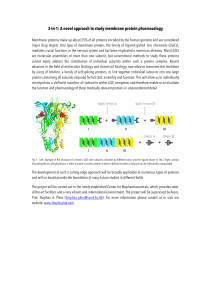
3-in-1: A novel approach to study membrane protein pharmacology
... mediates crucial functions in the nervous system and has been implicated a numerous diseases. Most LGICs are molecular assemblies of more than one subunit, but conventional methods to study these proteins cannot easily address the contribution of individual subunits within such a protein complex. Re ...
... mediates crucial functions in the nervous system and has been implicated a numerous diseases. Most LGICs are molecular assemblies of more than one subunit, but conventional methods to study these proteins cannot easily address the contribution of individual subunits within such a protein complex. Re ...
Review for Chapter 1
... How many electron pairs does carbon share in order to complete its valence shell? What are the most important functional groups that make of the molecules of life? What are they made of and what do they look like? ...
... How many electron pairs does carbon share in order to complete its valence shell? What are the most important functional groups that make of the molecules of life? What are they made of and what do they look like? ...
Moonlighting and pleiotropy among regulators of the degradation
... Multifunctional proteins are generally referred to as either pleiotropic or moonlighting proteins. Although define similarly, the meaning is these terms is quite different. While a moonlighting protein harbors several autonomous functions, pleiotropy refers to a single-functional protein, which infl ...
... Multifunctional proteins are generally referred to as either pleiotropic or moonlighting proteins. Although define similarly, the meaning is these terms is quite different. While a moonlighting protein harbors several autonomous functions, pleiotropy refers to a single-functional protein, which infl ...
5. Nucleotides are covalently linked to form nucleic acids by the
... triphosphate; positive; negative; zero; cellular respiration; photosynthesis; reduction 16. In biology, questions about the present are inescapably linked to questions about the past. Which one of the three tenets of the cell theory best clarifies this statement? ...
... triphosphate; positive; negative; zero; cellular respiration; photosynthesis; reduction 16. In biology, questions about the present are inescapably linked to questions about the past. Which one of the three tenets of the cell theory best clarifies this statement? ...
Title - Iowa State University
... 1.) Which of the following will NOT change a protein function: a.) Phosphorylation of a protein b.) Temperature of the environment c.) Sequence of the amino acid composition d.) pH of environment e.) None of the above 2.) The structural level of a protein in which alpha helixes and beta sheets form ...
... 1.) Which of the following will NOT change a protein function: a.) Phosphorylation of a protein b.) Temperature of the environment c.) Sequence of the amino acid composition d.) pH of environment e.) None of the above 2.) The structural level of a protein in which alpha helixes and beta sheets form ...
CALL FOR PAPERS 6th IEEE International Conference on
... Advances in high-throughput technologies such as DNA sequencing and mass spectrometry are profoundly transforming life sciences, resulting in the collection of unprecedented amounts of biological and medical data. Using this data to advance our knowledge about fundamental biological processes and im ...
... Advances in high-throughput technologies such as DNA sequencing and mass spectrometry are profoundly transforming life sciences, resulting in the collection of unprecedented amounts of biological and medical data. Using this data to advance our knowledge about fundamental biological processes and im ...
Coevolution in protein families: a functional correlation study.
... problem can be solved in a single step, without using any iterative scheme. We tested the efficiency and the prediction capacity of the method for intraprotein contacts on 131 proteins: we achieved the highest performance compared to all other methods. Concerning interprotein interactions, a specifi ...
... problem can be solved in a single step, without using any iterative scheme. We tested the efficiency and the prediction capacity of the method for intraprotein contacts on 131 proteins: we achieved the highest performance compared to all other methods. Concerning interprotein interactions, a specifi ...