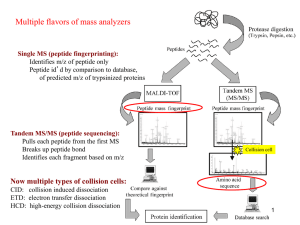
No Slide Title
... • Group of residues with high contact density, number of contacts within domains is higher than the number of contacts between domains. • A stable unit of protein structure that can fold autonomously • A rigid body linked to other domains by flexible linkers • A portion of the protein that can be ac ...
... • Group of residues with high contact density, number of contacts within domains is higher than the number of contacts between domains. • A stable unit of protein structure that can fold autonomously • A rigid body linked to other domains by flexible linkers • A portion of the protein that can be ac ...
Protein Ubiquitination
... ER Capacity: The protein concentration in the ER lumen is 100 mg/ml, it is essential that protein chaperones facilitate protein folding by preventing aggregation of protein folding intermediates and by correcting misfolded proteins. BiP/GRP78 – uses the energy from ATP hydrolysis to facilitate foldi ...
... ER Capacity: The protein concentration in the ER lumen is 100 mg/ml, it is essential that protein chaperones facilitate protein folding by preventing aggregation of protein folding intermediates and by correcting misfolded proteins. BiP/GRP78 – uses the energy from ATP hydrolysis to facilitate foldi ...
Datasheet for Protein Marker, Broad Range (2-212 kDa)
... Description: Protein Marker, Broad Range is a mixture of purified proteins with known amino acid sequences. They are resolved to 13 sharp bands when analyzed by SDS-PAGE (Tris-Glycine) and stained with Coomassie Blue R-250 (1). Two bands (BSA, MW 66.4 kDa and Triosephosphate isomerase, MW 27.0 kDa) ...
... Description: Protein Marker, Broad Range is a mixture of purified proteins with known amino acid sequences. They are resolved to 13 sharp bands when analyzed by SDS-PAGE (Tris-Glycine) and stained with Coomassie Blue R-250 (1). Two bands (BSA, MW 66.4 kDa and Triosephosphate isomerase, MW 27.0 kDa) ...
structures
... • Over long periods of time a sequence will acquire random mutations. – These mutations may result in a new amino acid at a given position, the deletion of an amino acid, or the introduction of a new one. – Over VERY long periods of time two sequences may diverge so much that their relationship can ...
... • Over long periods of time a sequence will acquire random mutations. – These mutations may result in a new amino acid at a given position, the deletion of an amino acid, or the introduction of a new one. – Over VERY long periods of time two sequences may diverge so much that their relationship can ...
Scoring Docked Protein Complexes with Hydrogen Bonds
... My work involves the scoring of these protein-protein complexes based on the number of hydrogen bonds present in the interface of the complex. This work was done on the basis that hydrogen bonds provide at least specificity to protein-protein complexes, if not stability.The code I wrote analyzes pro ...
... My work involves the scoring of these protein-protein complexes based on the number of hydrogen bonds present in the interface of the complex. This work was done on the basis that hydrogen bonds provide at least specificity to protein-protein complexes, if not stability.The code I wrote analyzes pro ...
Nitroshure-general info
... Nitroshure can be used to replace up to 1 to 2 lbs. of dietary ingredients, particularly protein supplements. This will allow for an additional 1 to 1.5 lb of DM from other non-protein dense ingredients such as forage, energy concentrates or byproducts to be used along with Nitroshure. This allows t ...
... Nitroshure can be used to replace up to 1 to 2 lbs. of dietary ingredients, particularly protein supplements. This will allow for an additional 1 to 1.5 lb of DM from other non-protein dense ingredients such as forage, energy concentrates or byproducts to be used along with Nitroshure. This allows t ...
Protein and its functional properties in food
... flour and albumin in egg white. Some types of protein help with reactions – these are called enzymes, whilst others form part of the structure of the cells. ...
... flour and albumin in egg white. Some types of protein help with reactions – these are called enzymes, whilst others form part of the structure of the cells. ...
THERAPUETIC DISCOVERY BY MODELLING
... consuming and expensive, with very low hit rates for the amount of resources expended. Computational screening of compounds against structures of protein targets offers a way to speed up discovery time and reduce costs, but such techniques have typically had low accuracy and need high resolution str ...
... consuming and expensive, with very low hit rates for the amount of resources expended. Computational screening of compounds against structures of protein targets offers a way to speed up discovery time and reduce costs, but such techniques have typically had low accuracy and need high resolution str ...
IOSR Journal of Pharmacy and Biological Sciences (IOSR-JPBS) e-ISSN: 2278-3008.
... Insulin is a peptide hormone, produced by beta cells of the pancreas, and is central to regulating carbohydrate and fat metabolism in the body. Insulin causes cells in the liver, skeletal muscles, and fat tissue to absorb glucose from the blood. In the liver and skeletal muscles, glucose is stored a ...
... Insulin is a peptide hormone, produced by beta cells of the pancreas, and is central to regulating carbohydrate and fat metabolism in the body. Insulin causes cells in the liver, skeletal muscles, and fat tissue to absorb glucose from the blood. In the liver and skeletal muscles, glucose is stored a ...
Protein – Protein Interactions
... A protein interaction, (P1, P2), is explained by a domain pair, (D1, D2), if P1 includes one domain and P2 includes the other. Find the minimum number of domain pairs that explains the databank. Equivalent to Minimum Set Cover problem. ...
... A protein interaction, (P1, P2), is explained by a domain pair, (D1, D2), if P1 includes one domain and P2 includes the other. Find the minimum number of domain pairs that explains the databank. Equivalent to Minimum Set Cover problem. ...
Genomics in Drug Discovery
... processes • Understanding protein functions and relationships is very important for drug design ...
... processes • Understanding protein functions and relationships is very important for drug design ...
Proceeding - ETH Zürich
... resistance is reduced. The random structure changes to a shear-oriented structure with progressively ...
... resistance is reduced. The random structure changes to a shear-oriented structure with progressively ...
Module 7 - Protein Structure Prediction
... is no homologue in the structural databases then things become rather more difficult, but not impossible. Even with no no homologues of known structure it may be possible to use fold recognition methods. There is a so called “twilight area” of 20-30% sequence identity, where it is difficult to asses ...
... is no homologue in the structural databases then things become rather more difficult, but not impossible. Even with no no homologues of known structure it may be possible to use fold recognition methods. There is a so called “twilight area” of 20-30% sequence identity, where it is difficult to asses ...
Proteomics2_2012
... - mixture model approach: take the distribution of ALL scores S - this is a mixture of ‘correct’ PSMs and ‘incorrect’ PSMs - but we don’t know which are correct or incorrect - scores from decoy comparison are included, which can provide some idea of the distribution of ‘incorrect’ scores -EM or Baye ...
... - mixture model approach: take the distribution of ALL scores S - this is a mixture of ‘correct’ PSMs and ‘incorrect’ PSMs - but we don’t know which are correct or incorrect - scores from decoy comparison are included, which can provide some idea of the distribution of ‘incorrect’ scores -EM or Baye ...
Ch.5
... If pH > pI, then the protein is negatively charged (acidic proteins have pI < 7) If pH < pI, the protein is positively charged (basic proteins have pI > 7) ...
... If pH > pI, then the protein is negatively charged (acidic proteins have pI < 7) If pH < pI, the protein is positively charged (basic proteins have pI > 7) ...
ANSWER: Proteins, Amino Acids and Carbs
... While this might be fine for the general population for whom soy beans are no more than an important foodstuff, this is not good enough for those using soy for the specific chemical makeup of its protein. It is very important that the soy proteins they use metabolize into the correct balance of amin ...
... While this might be fine for the general population for whom soy beans are no more than an important foodstuff, this is not good enough for those using soy for the specific chemical makeup of its protein. It is very important that the soy proteins they use metabolize into the correct balance of amin ...
Read more... - Barrhaven Business Improvement Area
... Ideal Protein company has spent a great deal of time and research making sure that they offer a wide variety of foods that taste great. “Many patients are pleasantly surprised at how good the products taste”. Unlike other weight loss programs, Ideal Protein is more than a product. In addition to usi ...
... Ideal Protein company has spent a great deal of time and research making sure that they offer a wide variety of foods that taste great. “Many patients are pleasantly surprised at how good the products taste”. Unlike other weight loss programs, Ideal Protein is more than a product. In addition to usi ...
biological process
... http://www.ebi.ac.uk/interpro/ ExPASy Proteomics Server The ExPASy (Expert Protein Analysis System) proteomics server of the Swiss Institute of Bioinformatics (SIB) is dedicated to the analysis of protein sequences and structures as well as 2-D PAGE (Disclaimer / References). ...
... http://www.ebi.ac.uk/interpro/ ExPASy Proteomics Server The ExPASy (Expert Protein Analysis System) proteomics server of the Swiss Institute of Bioinformatics (SIB) is dedicated to the analysis of protein sequences and structures as well as 2-D PAGE (Disclaimer / References). ...
Nutrition and Your Health
... – Yes, the amino acids sequence in animal protein is closer to human sequencing than plant. But, the closest is human flesh; would you want to eat that? – Does faster growth equate to better health? ...
... – Yes, the amino acids sequence in animal protein is closer to human sequencing than plant. But, the closest is human flesh; would you want to eat that? – Does faster growth equate to better health? ...
Protein Structure Prediction
... AlphaPred: A web server for prediction of -turns in proteins (http://www.imtech.res.in/raghava/alphapred/) Harpreet Kaur and G P S Raghava (2003) Prediction of -turns in proteins using PSI-BLAST profiles and secondary structure information. Proteins . ...
... AlphaPred: A web server for prediction of -turns in proteins (http://www.imtech.res.in/raghava/alphapred/) Harpreet Kaur and G P S Raghava (2003) Prediction of -turns in proteins using PSI-BLAST profiles and secondary structure information. Proteins . ...
Protein Quantification:
... By eye, what would you say the level of darkness is? Perhaps the value is between 3-4 on our scale. Essentially, what you have done is to create a standard curve in your mind and you compared the levels of darkness using your eye to qualitatively, assess the unknown. In biochemistry, we need to be m ...
... By eye, what would you say the level of darkness is? Perhaps the value is between 3-4 on our scale. Essentially, what you have done is to create a standard curve in your mind and you compared the levels of darkness using your eye to qualitatively, assess the unknown. In biochemistry, we need to be m ...
No Slide Title
... structure (conformation) z In turn, a protein’s structure determines the function of that protein ...
... structure (conformation) z In turn, a protein’s structure determines the function of that protein ...
View/Download
... And NuAquos, from New Whey Nutrition, contains a propriety blend of protein, electrolytes, vitamins and minerals. With 12 grams of protein, it promotes recovery and rehydration. “We think one of the problems protein’s had in the past is they haven’t been able to make a good tasting product,” said Ch ...
... And NuAquos, from New Whey Nutrition, contains a propriety blend of protein, electrolytes, vitamins and minerals. With 12 grams of protein, it promotes recovery and rehydration. “We think one of the problems protein’s had in the past is they haven’t been able to make a good tasting product,” said Ch ...
protein
... 1. Name the elemental composition of protein. 2. What is meant by high biological value protein? 3. What is meant by low biological value protein? 4. What are complementary protein? Give examples. 5. Give three functions of protein. Give examples. 6. What is denaturation? ...
... 1. Name the elemental composition of protein. 2. What is meant by high biological value protein? 3. What is meant by low biological value protein? 4. What are complementary protein? Give examples. 5. Give three functions of protein. Give examples. 6. What is denaturation? ...
Heterologous expression and purification of proteins in E. coli
... – permits a lot of different buffer conditions BUT – GF columns are expensive and fragile – sample size should be very small (<1% of column volume) – not a very good „first step“, more suitable for final refinement ...
... – permits a lot of different buffer conditions BUT – GF columns are expensive and fragile – sample size should be very small (<1% of column volume) – not a very good „first step“, more suitable for final refinement ...
Rosetta@home

Rosetta@home is a distributed computing project for protein structure prediction on the Berkeley Open Infrastructure for Network Computing (BOINC) platform, run by the Baker laboratory at the University of Washington. Rosetta@home aims to predict protein–protein docking and design new proteins with the help of about sixty thousand active volunteered computers processing at 83 teraFLOPS on average as of April 18, 2014. Foldit, a Rosetta@Home videogame, aims to reach these goals with a crowdsourcing approach. Though much of the project is oriented towards basic research on improving the accuracy and robustness of the proteomics methods, Rosetta@home also does applied research on malaria, Alzheimer's disease and other pathologies.Like all BOINC projects, Rosetta@home uses idle computer processing resources from volunteers' computers to perform calculations on individual workunits. Completed results are sent to a central project server where they are validated and assimilated into project databases. The project is cross-platform, and runs on a wide variety of hardware configurations. Users can view the progress of their individual protein structure prediction on the Rosetta@home screensaver.In addition to disease-related research, the Rosetta@home network serves as a testing framework for new methods in structural bioinformatics. These new methods are then used in other Rosetta-based applications, like RosettaDock and the Human Proteome Folding Project, after being sufficiently developed and proven stable on Rosetta@home's large and diverse collection of volunteer computers. Two particularly important tests for the new methods developed in Rosetta@home are the Critical Assessment of Techniques for Protein Structure Prediction (CASP) and Critical Assessment of Prediction of Interactions (CAPRI) experiments, biannual experiments which evaluate the state of the art in protein structure prediction and protein–protein docking prediction, respectively. Rosetta@home consistently ranks among the foremost docking predictors, and is one of the best tertiary structure predictors available.