Protein Targeting
... The life of protein Determined by N terminal amino acid Proteins with ala, met, gly, ser, val, thr etc at the N terminus have more half life Proteins with glu, gln, asp and asn have less half life The tagged proteins are turned over by a 26s protease complex. It leaves ubiquitin unaffected. ...
... The life of protein Determined by N terminal amino acid Proteins with ala, met, gly, ser, val, thr etc at the N terminus have more half life Proteins with glu, gln, asp and asn have less half life The tagged proteins are turned over by a 26s protease complex. It leaves ubiquitin unaffected. ...
The Structure and Function of Macromolecules
... There are many thousands of different types of enzymes alone – each specifically designed for a particular chemical reaction. ...
... There are many thousands of different types of enzymes alone – each specifically designed for a particular chemical reaction. ...
Tutorial 6
... 10) Measuring the speed of light for yourself. The accepted value for the speed of light is 2.99792458 x108 m/s. Measuring the speed of electromagnetic radiation or light normally requires delicate experiments involving expensive equipment. However, you can actually find a value for the speed of lig ...
... 10) Measuring the speed of light for yourself. The accepted value for the speed of light is 2.99792458 x108 m/s. Measuring the speed of electromagnetic radiation or light normally requires delicate experiments involving expensive equipment. However, you can actually find a value for the speed of lig ...
Amino Acids
... • Polar and charged residues tend to be on surface of protein, exposed to water, while hydrophobic residues tend to be buried ...
... • Polar and charged residues tend to be on surface of protein, exposed to water, while hydrophobic residues tend to be buried ...
Spectroscopy questions for midterm or semi-final exam, 2016
... energy. In XPS the photon is absorbed by an atom in a molecule, leading to ionization and the emission of a core (inner shell) electron. In UPS the photon interacts with valence levels of the molecule, leading to ionisation by removal of one of these valence electrons. 6. Apply the energy conservati ...
... energy. In XPS the photon is absorbed by an atom in a molecule, leading to ionization and the emission of a core (inner shell) electron. In UPS the photon interacts with valence levels of the molecule, leading to ionisation by removal of one of these valence electrons. 6. Apply the energy conservati ...
tutorial 6
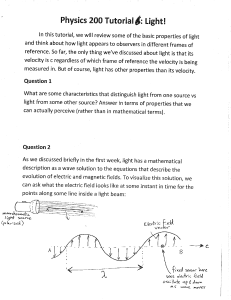
... at some instant in time. There are some places where the electric field points up and some places where the electric field points down (at any given time). The distance between two successive ,,peaks,, (places where the electric field is maximum) in the picture is called the WAVELENGTH, and is usual ...
... at some instant in time. There are some places where the electric field points up and some places where the electric field points down (at any given time). The distance between two successive ,,peaks,, (places where the electric field is maximum) in the picture is called the WAVELENGTH, and is usual ...
Science Jeopardy - Broward County Public Schools
... A molecule that forms much of a cell’s membrane (the outer layer). ...
... A molecule that forms much of a cell’s membrane (the outer layer). ...
Document
... protein, usually causing loss of function. -may involve complete unfolding -caused by changes in the protein’s environment -pH -temperature -salt concentration ...
... protein, usually causing loss of function. -may involve complete unfolding -caused by changes in the protein’s environment -pH -temperature -salt concentration ...
Proteins - Mr Waring`s Biology Blog
... on the inside, while the hydrophilic groups are on the outside. This makes many globular proteins soluble in water. ...
... on the inside, while the hydrophilic groups are on the outside. This makes many globular proteins soluble in water. ...
PS 250
... 2. Find the magnitude of the electric field at (2, 3) due to a charge of 3.0 nC at (2,-2) and a charge of –5.0 nC at (-2,0), where (x,y) denote x and y coordinates in meters. a. 1.4 N/C * b. 1.0 N/C c. 2.1 N/C d. 1.8 N/C e. 2.8 N/C 3. An electron and a proton are separated by 5.31 × 10 −11 m . What ...
... 2. Find the magnitude of the electric field at (2, 3) due to a charge of 3.0 nC at (2,-2) and a charge of –5.0 nC at (-2,0), where (x,y) denote x and y coordinates in meters. a. 1.4 N/C * b. 1.0 N/C c. 2.1 N/C d. 1.8 N/C e. 2.8 N/C 3. An electron and a proton are separated by 5.31 × 10 −11 m . What ...
New NMR experimental techniques: Protein structural compactness
... variety of binding partners. We propose taking advantage on the different relaxation dynamics of IDPs and globular proteins to understand the structural compactness and solvent accessibility by new NMR experiments based on a set of selective measurements of T1 relaxation delays (I). NMR spectroscopy ...
... variety of binding partners. We propose taking advantage on the different relaxation dynamics of IDPs and globular proteins to understand the structural compactness and solvent accessibility by new NMR experiments based on a set of selective measurements of T1 relaxation delays (I). NMR spectroscopy ...
Topic 2.2: Proteins
... The primary structure ensures that R groups are always in the same position, therefore the bonding between Rgroups will always be the same, and the hydrophobic and hydrophillic interactions will always be the same and therefore the tertialy structure of a specific protein is always identical. ...
... The primary structure ensures that R groups are always in the same position, therefore the bonding between Rgroups will always be the same, and the hydrophobic and hydrophillic interactions will always be the same and therefore the tertialy structure of a specific protein is always identical. ...
PROTEIN STRUCTURE SIMILARITY CALCULATION AND VISUALIZATION
... Input to moduleB or visualization module and the output The all by all pairwise similarity calculated in moduleA will be used as input to moduleB. Output should be connectivity graph (as shown in next slide) between all proteins. Each edge must display the similarity value. Preferred output will be ...
... Input to moduleB or visualization module and the output The all by all pairwise similarity calculated in moduleA will be used as input to moduleB. Output should be connectivity graph (as shown in next slide) between all proteins. Each edge must display the similarity value. Preferred output will be ...
Circular dichroism

Circular dichroism (CD) is dichroism involving circularly polarized light, i.e., the differential absorption of left- and right-handed light. Left-hand circular (LHC) and right-hand circular (RHC) polarized light represent two possible spin angular momentum states for a photon, and so circular dichroism is also referred to as dichroism for spin angular momentum. This phenomenon was discovered by Jean-Baptiste Biot, Augustin Fresnel, and Aimé Cotton in the first half of the 19th century. It is exhibited in the absorption bands of optically active chiral molecules. CD spectroscopy has a wide range of applications in many different fields. Most notably, UV CD is used to investigate the secondary structure of proteins. UV/Vis CD is used to investigate charge-transfer transitions. Near-infrared CD is used to investigate geometric and electronic structure by probing metal d→d transitions. Vibrational circular dichroism, which uses light from the infrared energy region, is used for structural studies of small organic molecules, and most recently proteins and DNA.