Chapter 15. Recognition of foreign molecules by the immune system
... All immunoglobulin domains have similar three-dimensional structures – Immunoglobulin fold ...
... All immunoglobulin domains have similar three-dimensional structures – Immunoglobulin fold ...
STUDIES ON SURFACE PROTEINS OF
... prove useful as a biochemi~al tool for classification of Leishmaniastrains. However, ,investigations on a large number of strains that have also been characterized by other parameters are needed to clarify this possibility. The presence of cross-reacting surface antigens suggests that it may be poss ...
... prove useful as a biochemi~al tool for classification of Leishmaniastrains. However, ,investigations on a large number of strains that have also been characterized by other parameters are needed to clarify this possibility. The presence of cross-reacting surface antigens suggests that it may be poss ...
Macromolecules
... Note that no break in the polymer chain (disruption of primary structure) is involved in denaturation. Denaturing agents disrupt stabilizing factors. Agents that disrupt hydrogen bonding: Heat -- thermal agitation (vibration, etc.) -- will denature proteins or nucleic acids. Heat denaturation of DNA ...
... Note that no break in the polymer chain (disruption of primary structure) is involved in denaturation. Denaturing agents disrupt stabilizing factors. Agents that disrupt hydrogen bonding: Heat -- thermal agitation (vibration, etc.) -- will denature proteins or nucleic acids. Heat denaturation of DNA ...
Membranlar - mustafaaltinisik.org.uk
... hydrophilic residues • Found in outer membrane of Gm- bacteria & mitochondria eg maltoporin ...
... hydrophilic residues • Found in outer membrane of Gm- bacteria & mitochondria eg maltoporin ...
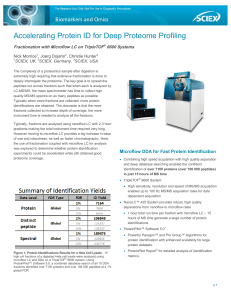
Accelerating Protein ID for Deep Proteome Profiling
... can be reflected in the score from the subsequent database search. Here, the score distribution (Figure 4, top) indicated that the majority of MS/MS spectra acquired were of high quality, score of 8 or more (blue). When the distribution for the MS/MS spectra that resulted in a confident peptide ID i ...
... can be reflected in the score from the subsequent database search. Here, the score distribution (Figure 4, top) indicated that the majority of MS/MS spectra acquired were of high quality, score of 8 or more (blue). When the distribution for the MS/MS spectra that resulted in a confident peptide ID i ...
Slide 1
... •Let’s predict the secondary structure of the little transmembrane protein using a multiple sequence alignment with homologs. •Load littleMSA_fasta.txt on JalView •Calculate secondary structure prediction using Web Service > Secondary Structure Prediction > Jnet (Do not select any sequences when doi ...
... •Let’s predict the secondary structure of the little transmembrane protein using a multiple sequence alignment with homologs. •Load littleMSA_fasta.txt on JalView •Calculate secondary structure prediction using Web Service > Secondary Structure Prediction > Jnet (Do not select any sequences when doi ...
- Wiley Online Library
... assays. Our results showed that the maize protein MA16 is an RNA-binding protein that binds preferentially to uridine and guanosine-rich RNAs (Ludevid et al., 1992). Regulation studies indicated that MA16 mRNA had a basal level of expression in several tissues including embryos and seedlings. We als ...
... assays. Our results showed that the maize protein MA16 is an RNA-binding protein that binds preferentially to uridine and guanosine-rich RNAs (Ludevid et al., 1992). Regulation studies indicated that MA16 mRNA had a basal level of expression in several tissues including embryos and seedlings. We als ...
Good Nutrition for Dancers
... generally lose up to two litres of water each day in perspiration and moisture loss from basic body functions. It is important to consume fluid throughout the day and not wait until you feel thirsty—a sign that it is too late! The amount of water needed varies between people. One way to calculate you ...
... generally lose up to two litres of water each day in perspiration and moisture loss from basic body functions. It is important to consume fluid throughout the day and not wait until you feel thirsty—a sign that it is too late! The amount of water needed varies between people. One way to calculate you ...
Supplementary Information
... Figure 6: P53 activation (phosphorylation of serine 15) and p53-target gene expression after Cyclosporin A (CsA) treatment and UVC irradiation. H1299(p53-/-), H1299(p5372P) and H1299(p5372R) cells were cultivated at 32oC and treated with CsA (0.5 µM) or UVC irradiation (25 J/m2) for four hours, res ...
... Figure 6: P53 activation (phosphorylation of serine 15) and p53-target gene expression after Cyclosporin A (CsA) treatment and UVC irradiation. H1299(p53-/-), H1299(p5372P) and H1299(p5372R) cells were cultivated at 32oC and treated with CsA (0.5 µM) or UVC irradiation (25 J/m2) for four hours, res ...
The PYRIN domain: a novel motif found in apoptosis and
... here that the PYRIN domain is a novel protein module that likely mediates protein-protein interactions in apoptotic and inflammatory signaling pathways. CARD4 (also called NOD1) is a CED-4/Apaf-1 family member that activates NF-kB signaling and induces apoptosis.3 ± 5 CARD4 contains three putative f ...
... here that the PYRIN domain is a novel protein module that likely mediates protein-protein interactions in apoptotic and inflammatory signaling pathways. CARD4 (also called NOD1) is a CED-4/Apaf-1 family member that activates NF-kB signaling and induces apoptosis.3 ± 5 CARD4 contains three putative f ...
Proteins
... signals from one body system to another insulin movement muscle immune system protect against germs enzymes Regents Biology help chemical reactions ...
... signals from one body system to another insulin movement muscle immune system protect against germs enzymes Regents Biology help chemical reactions ...
- BioTek Instruments
... absorptivities cannot be known. Typically a calibration curve using a model protein, such as bovine serum albumin (BSA) is used to interpolate sample protein mixture concentrations from OD measurements. There will of course be some inaccuracy in this method, the extent determined by the relative exp ...
... absorptivities cannot be known. Typically a calibration curve using a model protein, such as bovine serum albumin (BSA) is used to interpolate sample protein mixture concentrations from OD measurements. There will of course be some inaccuracy in this method, the extent determined by the relative exp ...
English
... 1. Amino acids contain carbon, hydrogen, and oxygen. In plants, proteins are largely concentrated in the growing portions, especially leaves and seeds. 2. Essential amino acids cannot be produced fast enough by animals, and therefore must be supplied in the livestock feed. Maize and cotton seed cake ...
... 1. Amino acids contain carbon, hydrogen, and oxygen. In plants, proteins are largely concentrated in the growing portions, especially leaves and seeds. 2. Essential amino acids cannot be produced fast enough by animals, and therefore must be supplied in the livestock feed. Maize and cotton seed cake ...
yeast - chem.uwec.edu
... Create the Bait Plasmid Construct from the gene of interest and the DNA binding domain of Gal4 or LexA or other suitable domain Transform with the bait construct a yeast strain lacking the promoter for the reporter genes and select for transformed yeast Transform the yeast again with the library ...
... Create the Bait Plasmid Construct from the gene of interest and the DNA binding domain of Gal4 or LexA or other suitable domain Transform with the bait construct a yeast strain lacking the promoter for the reporter genes and select for transformed yeast Transform the yeast again with the library ...
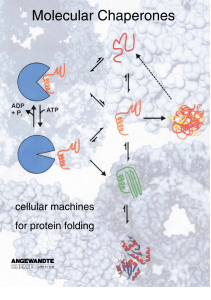
Molecular Chaperones - Cellular Machines for Protein Folding
... clearly showed that the three-dimensional structure of this protein is encoded in its amino acid sequence, and that no other factors are required for structure acquisition. Thus, protein folding is an autonomous and, given the proper conditions, spontaneous process. Following the work of Anfinsen an ...
... clearly showed that the three-dimensional structure of this protein is encoded in its amino acid sequence, and that no other factors are required for structure acquisition. Thus, protein folding is an autonomous and, given the proper conditions, spontaneous process. Following the work of Anfinsen an ...
The Quiet Revolution! - Montana State University
... • March Women learns to recognize kwashiorkor and make intervention diet for at-risk children • Sept women discourage children’s grasshopper hunting and eating due to pesticides in nearby cotton fields • Sept students and mentors introduce brick wall concept of complete proteins ...
... • March Women learns to recognize kwashiorkor and make intervention diet for at-risk children • Sept women discourage children’s grasshopper hunting and eating due to pesticides in nearby cotton fields • Sept students and mentors introduce brick wall concept of complete proteins ...
Chemistry 160 Exam 2 Key Pg. Chemistry 160 Exam 2 key Please
... (10) 6. Describe the 4 levels of protein structure. Primary = amino acid sequence Secondary = simple folding like α helix or beta sheet Supersecondary = patterns of secondary Tertiary = 3 D structure Quaternary = subunit interactions (5) 7. Give two types of forces that hold proteins in their 3-D sh ...
... (10) 6. Describe the 4 levels of protein structure. Primary = amino acid sequence Secondary = simple folding like α helix or beta sheet Supersecondary = patterns of secondary Tertiary = 3 D structure Quaternary = subunit interactions (5) 7. Give two types of forces that hold proteins in their 3-D sh ...
Homology Modeling Zinc Fingers – Introduction zf
... most of the residues reach an equilibrium point that is highly similar to the crystal structure. Cluster analysis revealed that the cluster with the most amount of neighbors is in general highly similar to the crystal structure. There are a few residues that are seen in the simulation that seem to f ...
... most of the residues reach an equilibrium point that is highly similar to the crystal structure. Cluster analysis revealed that the cluster with the most amount of neighbors is in general highly similar to the crystal structure. There are a few residues that are seen in the simulation that seem to f ...
Domain structure and sequence similarities in cartilage proteoglycan
... The large, aggregating chondroitin sulphate proteoglycan of cartilage, or aggrecan, is the predominant proteoglycan in cartilage. It has a monomeric relative M , of approx. 2.4 x lo", is highly substituted with chondroitin sulphate and keratan sulphate glycosaminoglycans, as well as N- and 0linked o ...
... The large, aggregating chondroitin sulphate proteoglycan of cartilage, or aggrecan, is the predominant proteoglycan in cartilage. It has a monomeric relative M , of approx. 2.4 x lo", is highly substituted with chondroitin sulphate and keratan sulphate glycosaminoglycans, as well as N- and 0linked o ...
Protein - HCC Learning Web
... 100 amino acids. The monomer units in the chain are known as amino acid residues. The average protein contains about 350 amino acid residues although proteins with as many as 1000 residues and those with as few as 100 are not uncommon. The sequence or order of amino acids along a polypeptide chain i ...
... 100 amino acids. The monomer units in the chain are known as amino acid residues. The average protein contains about 350 amino acid residues although proteins with as many as 1000 residues and those with as few as 100 are not uncommon. The sequence or order of amino acids along a polypeptide chain i ...
Materials and Methods - UROP
... Integrating nanotechnology with biology could allow the detailed study of biological systems at the single molecule level. For example, rather than studying the dynamics of proteins in the traditional, ensemble manner, nanotechnology could uncover how an individual protein interacts with its surroun ...
... Integrating nanotechnology with biology could allow the detailed study of biological systems at the single molecule level. For example, rather than studying the dynamics of proteins in the traditional, ensemble manner, nanotechnology could uncover how an individual protein interacts with its surroun ...
BIOL562_Lecture_13
... Protein folding Four levels of protein structure; need correct tertiary structure to be activated; a dynamic process; for large proteins, renaturation is not always spontaneous due to (1) tendency to form insoluable aggregates; (2) more stable alternative folding pathways. Figure 13.25-26 Genomes 3 ...
... Protein folding Four levels of protein structure; need correct tertiary structure to be activated; a dynamic process; for large proteins, renaturation is not always spontaneous due to (1) tendency to form insoluable aggregates; (2) more stable alternative folding pathways. Figure 13.25-26 Genomes 3 ...
The database of epoxide hydrolases and
... the α/β hydrolase fold are well conserved. The cap domains are also similar in shape and size, and consist of 4–5 α-helices arranged in 2 layers. The classification of EHs based on the length of NC- and cap-loop (Barth et al., 2004) was applied to 11 homologous families. Cluster I includes soluble m ...
... the α/β hydrolase fold are well conserved. The cap domains are also similar in shape and size, and consist of 4–5 α-helices arranged in 2 layers. The classification of EHs based on the length of NC- and cap-loop (Barth et al., 2004) was applied to 11 homologous families. Cluster I includes soluble m ...
A High Yield Method for the Removal of Detergents from Low
... While mass spectrometry (MS) occupies a central role in high-throughput proteomic analysis, sample preparation ahead of MS analysis is pivotal to successful protein characterization. Sample complexity, interfering substances and large dynamic range of protein and peptide concentrations are major hur ...
... While mass spectrometry (MS) occupies a central role in high-throughput proteomic analysis, sample preparation ahead of MS analysis is pivotal to successful protein characterization. Sample complexity, interfering substances and large dynamic range of protein and peptide concentrations are major hur ...
Protein folding

Protein folding is the process by which a protein structure assumes its functional shape or conformation. It is the physical process by which a polypeptide folds into its characteristic and functional three-dimensional structure from random coil.Each protein exists as an unfolded polypeptide or random coil when translated from a sequence of mRNA to a linear chain of amino acids. This polypeptide lacks any stable (long-lasting) three-dimensional structure (the left hand side of the first figure). Amino acids interact with each other to produce a well-defined three-dimensional structure, the folded protein (the right hand side of the figure), known as the native state. The resulting three-dimensional structure is determined by the amino acid sequence (Anfinsen's dogma). Experiments beginning in the 1980s indicate the codon for an amino acid can also influence protein structure.The correct three-dimensional structure is essential to function, although some parts of functional proteins may remain unfolded, so that protein dynamics is important. Failure to fold into native structure generally produces inactive proteins, but in some instances misfolded proteins have modified or toxic functionality. Several neurodegenerative and other diseases are believed to result from the accumulation of amyloid fibrils formed by misfolded proteins. Many allergies are caused by incorrect folding of some proteins, because the immune system does not produce antibodies for certain protein structures.