200 THINGS TO KNOW AP Biology TEST
... PGA and PGAL C4 plants outcompete C3 in high sunlight because they store extra Carbon to use when CO2 conc low. CAM plants are desert succulents ( cactus) stomata close during day open at night to avoid dessication) ( C3 and C4 plants stomata opened up during day) 91. C4 uses 5ATP per molecule C3 on ...
... PGA and PGAL C4 plants outcompete C3 in high sunlight because they store extra Carbon to use when CO2 conc low. CAM plants are desert succulents ( cactus) stomata close during day open at night to avoid dessication) ( C3 and C4 plants stomata opened up during day) 91. C4 uses 5ATP per molecule C3 on ...
Genetic Engineering
... Electrophoresis 1. “Cut” DNA sample with restriction enzymes. 2. Run the DNA fragments through a gel. ...
... Electrophoresis 1. “Cut” DNA sample with restriction enzymes. 2. Run the DNA fragments through a gel. ...
Science Notebook DNA, RNA, and Protein
... a group of three nitrogenous bases in DNA or mRNA that code for one amino acid nucleic acid made of ribose, phosphate, and one of four nitrogenous bases—adenine, cytosine, guanine, or uracil intervening DNA sequences that are transcribed and then removed from the final mRNA process by which mRNA dir ...
... a group of three nitrogenous bases in DNA or mRNA that code for one amino acid nucleic acid made of ribose, phosphate, and one of four nitrogenous bases—adenine, cytosine, guanine, or uracil intervening DNA sequences that are transcribed and then removed from the final mRNA process by which mRNA dir ...
11/01 Molecular genetic analysis and biotechnology
... DNA via a bacteria cell • Cloning vector: a replicating DNA molecule attached with a foreign DNA fragment to be introduced into a cell – Has features that make it easier to insert DNA and select for presence of vector in cell. • Origin of replication • Antibiotic resistance gene • Cloning site ...
... DNA via a bacteria cell • Cloning vector: a replicating DNA molecule attached with a foreign DNA fragment to be introduced into a cell – Has features that make it easier to insert DNA and select for presence of vector in cell. • Origin of replication • Antibiotic resistance gene • Cloning site ...
Microbial Genetics
... • The genotype of an individual refers to its particular genetic make-up. It’s the potential for all characteristics we can observe. A particular gene may have some variation in the sequence between individuals, called alleles. • The phenotype is the actual characteristics observed for an individua ...
... • The genotype of an individual refers to its particular genetic make-up. It’s the potential for all characteristics we can observe. A particular gene may have some variation in the sequence between individuals, called alleles. • The phenotype is the actual characteristics observed for an individua ...
FREE Sample Here - We can offer most test bank and
... especially if internal organs such as the liver, lungs, or kidneys have been damaged. One method of treatment involves injecting a blood-clotting factor that has been purified from blood donations. This factor is a protein encoded by a human gene. Suggest a way in which modern genetic technology cou ...
... especially if internal organs such as the liver, lungs, or kidneys have been damaged. One method of treatment involves injecting a blood-clotting factor that has been purified from blood donations. This factor is a protein encoded by a human gene. Suggest a way in which modern genetic technology cou ...
Ch. 10 Vocabs
... -Replication fork: a Y-shaped point that results when the two strands of a DNA double helix separate so that the DNA molecule can be replicated. -DNA polymerase: an enzyme that catalyzes the formation of the DNA molecule. -Semi-conservative replication: in each new DNA double helix, one strand is fr ...
... -Replication fork: a Y-shaped point that results when the two strands of a DNA double helix separate so that the DNA molecule can be replicated. -DNA polymerase: an enzyme that catalyzes the formation of the DNA molecule. -Semi-conservative replication: in each new DNA double helix, one strand is fr ...
Enzyme Induction
... • Primers define the region to be amplified • Short single-stranded DNAs • In PCR, always used in pairs • You choose their sequences • Primers will hybridize to complementary sequences in the target DNA to be amplified • Taq polymerase synthesizes new DNA onto the 3’ end of each primer • Whatever DN ...
... • Primers define the region to be amplified • Short single-stranded DNAs • In PCR, always used in pairs • You choose their sequences • Primers will hybridize to complementary sequences in the target DNA to be amplified • Taq polymerase synthesizes new DNA onto the 3’ end of each primer • Whatever DN ...
Biotechnology and Genetic Engineering
... Sequences examined in DNA fingerprinting • VNTRs-variable number tandem repeats; composed of 8-80 bp repeat units (e.g., [GCGCAATG]n) which are tandemly repeated so that the overall length is 1-30 kb • STRs-short tandem repeats; composed of 2-7 bp repeat units (e.g., [AC]n) which are tandemly repea ...
... Sequences examined in DNA fingerprinting • VNTRs-variable number tandem repeats; composed of 8-80 bp repeat units (e.g., [GCGCAATG]n) which are tandemly repeated so that the overall length is 1-30 kb • STRs-short tandem repeats; composed of 2-7 bp repeat units (e.g., [AC]n) which are tandemly repea ...
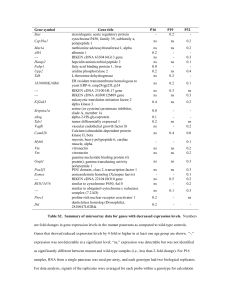
Table S2. Summary of microarray data for genes with decreased
... Table S2. Summary of microarray data for genes with decreased expression levels. Numbers are fold changes in gene expression levels in the mutant pancreata as compared to wild-type controls. Genes that showed reduced expression levels by 4 fold or higher in at least one age group are shown. “-,” exp ...
... Table S2. Summary of microarray data for genes with decreased expression levels. Numbers are fold changes in gene expression levels in the mutant pancreata as compared to wild-type controls. Genes that showed reduced expression levels by 4 fold or higher in at least one age group are shown. “-,” exp ...
Gene Expression Gene expression involves coded information on
... acids. The anticodon on the tRNA matchs up with the complementary codon on the mRNA in the ribosome. The amino acid is ‘dropped off’ and a peptide bond forms between amino acids. The anticondon then goes back into the cytoplasm to attach to a specific amino acid and match up with another complement ...
... acids. The anticodon on the tRNA matchs up with the complementary codon on the mRNA in the ribosome. The amino acid is ‘dropped off’ and a peptide bond forms between amino acids. The anticondon then goes back into the cytoplasm to attach to a specific amino acid and match up with another complement ...
Learning Targets - Unit 9 DNA, RNA, Proteins, Mutation
... Your goal for the end of this unit is to be able to say, “I ...
... Your goal for the end of this unit is to be able to say, “I ...
DNA Transcription and Translation
... nucleus so that the cell can make a protein out of the information obtained from the DNA in the gene Every 3 nitrogen bases in the DNA that makes up a gene is called a codon, and codes for a specific amino acid DNA does not leave the nucleus so a carrier molecule called messanger RNA (mRNA) i ...
... nucleus so that the cell can make a protein out of the information obtained from the DNA in the gene Every 3 nitrogen bases in the DNA that makes up a gene is called a codon, and codes for a specific amino acid DNA does not leave the nucleus so a carrier molecule called messanger RNA (mRNA) i ...
Regulation of Gene Expression in Eukaryotes
... Opportunities for the control of gene expression in the eukaryotic cell ...
... Opportunities for the control of gene expression in the eukaryotic cell ...
Genetic engineering and biotechnology techniques
... genome mapping before cheaper methods such as PCR (polymerase chain reaction) and DNA sequencing came along ...
... genome mapping before cheaper methods such as PCR (polymerase chain reaction) and DNA sequencing came along ...
Biology 303 EXAM II 3/14/00 NAME
... A. To allow resistant transformants to grow in selective medium B. To distinguish introns from exons C. To screen for vectors with inserts D. To allow plasmid replication ...
... A. To allow resistant transformants to grow in selective medium B. To distinguish introns from exons C. To screen for vectors with inserts D. To allow plasmid replication ...
Real-time polymerase chain reaction

A real-time polymerase chain reaction is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR, i.e. in real-time, and not at its end, as in conventional PCR. Real-time PCR can be used quantitatively (Quantitative real-time PCR), semi-quantitatively, i.e. above/below a certain amount of DNA molecules (Semi quantitative real-time PCR) or qualitatively (Qualitative real-time PCR).Two common methods for the detection of PCR products in real-time PCR are: (1) non-specific fluorescent dyes that intercalate with any double-stranded DNA, and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter which permits detection only after hybridization of the probe with its complementary sequence.The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines propose that the abbreviation qPCR be used for quantitative real-time PCR and that RT-qPCR be used for reverse transcription–qPCR [1]. The acronym ""RT-PCR"" commonly denotes reverse transcription polymerase chain reaction and not real-time PCR, but not all authors adhere to this convention.