Biomolecules in water and water in biomolecules
... We have been developing a new theory for the molecular recognition by protein based on the statistical mechanics of liquids, or the 3D-RISM/RISM theory. The theory has demonstrated its amazing capability of “predicting” the process from the frist principle. [1] However, what we have investigated so ...
... We have been developing a new theory for the molecular recognition by protein based on the statistical mechanics of liquids, or the 3D-RISM/RISM theory. The theory has demonstrated its amazing capability of “predicting” the process from the frist principle. [1] However, what we have investigated so ...
Dr. Fernando L. Barroso da Silva Protein complexation driven by
... technology, pharmaceutical formulations, medical and bioprocess technology. The oftenobserved strong dependency of such complexes formation on salt and pH indicates that electrostatic interactions are their main driving forces. It is clear that the interplay between the (bio)colloidal characteristic ...
... technology, pharmaceutical formulations, medical and bioprocess technology. The oftenobserved strong dependency of such complexes formation on salt and pH indicates that electrostatic interactions are their main driving forces. It is clear that the interplay between the (bio)colloidal characteristic ...
ppt
... Limitations of current databases: Do not contain strict/detailed descriptions of protocol (buffers, sample volume, staining techniques all important information for gel comparisons). Designed as 2D (and not proteomics) databases and therefore not readily expandable to incorporate other proteomics da ...
... Limitations of current databases: Do not contain strict/detailed descriptions of protocol (buffers, sample volume, staining techniques all important information for gel comparisons). Designed as 2D (and not proteomics) databases and therefore not readily expandable to incorporate other proteomics da ...
ASMS 2013 phosphopeptide poster
... dividing or activated. The macrophage is a cell that is waiting to be activated by external stimuli, therefore, unless the cell is exposed to activating factors, we expect that many (if not majority) of the signaling pathways based on phosphorylation or de-phosphorylation are at the lowest activity. ...
... dividing or activated. The macrophage is a cell that is waiting to be activated by external stimuli, therefore, unless the cell is exposed to activating factors, we expect that many (if not majority) of the signaling pathways based on phosphorylation or de-phosphorylation are at the lowest activity. ...
Coomassie Blue R-250 (SureStain)
... used in place of the R-250 form to create a rapid and convenient staining procedure. This capability of G-250 is due to its particular properties. Coomassie G-250 manifests a leuco form below pH 2. Solutions of the dye, dark blue black at pH 7, turn a clear tan upon acidification. The leuco form rec ...
... used in place of the R-250 form to create a rapid and convenient staining procedure. This capability of G-250 is due to its particular properties. Coomassie G-250 manifests a leuco form below pH 2. Solutions of the dye, dark blue black at pH 7, turn a clear tan upon acidification. The leuco form rec ...
OUTLINE
... Mass Spectrometry-1 the most recent and sensitive technique... • Mass Spectrometry is a method for determining the mass of molecules by producing and analyzing charged species (ions) • In the late 1980s, “soft” ionization techniques allows the production of ions of low energy and facilitated the io ...
... Mass Spectrometry-1 the most recent and sensitive technique... • Mass Spectrometry is a method for determining the mass of molecules by producing and analyzing charged species (ions) • In the late 1980s, “soft” ionization techniques allows the production of ions of low energy and facilitated the io ...
aminoacyl-tRNA
... targeting certain proteins to lysosomes • A 14-residue core oligosaccharide is built up by sequential addition of monosaccharide units to a dolichol-phosphate, which occurs on the cytosolic and lumen surfaces of the ER membrane. • The core is then enzymatically transferred en bloc to the side chain ...
... targeting certain proteins to lysosomes • A 14-residue core oligosaccharide is built up by sequential addition of monosaccharide units to a dolichol-phosphate, which occurs on the cytosolic and lumen surfaces of the ER membrane. • The core is then enzymatically transferred en bloc to the side chain ...
LS1a Fall 09
... Lane 1 provides evidence that the in vitro translation system is working as evidenced by the presence of Protein X expression when mRNA is added. a. What are some components that must be included in order to conduct in vitro translation in a test tube rather than in a cell? Ribosomes (large and sm ...
... Lane 1 provides evidence that the in vitro translation system is working as evidenced by the presence of Protein X expression when mRNA is added. a. What are some components that must be included in order to conduct in vitro translation in a test tube rather than in a cell? Ribosomes (large and sm ...
mcnair 2003 poster template
... pipetted off. The isolated protein can then be eluted for use in experiments. For this experiment, the goal is to isolate the entire complex of proteins associated with dynein during mitosis. If augmin is among these proteins, it suggests an interaction between augmin and dynein. ...
... pipetted off. The isolated protein can then be eluted for use in experiments. For this experiment, the goal is to isolate the entire complex of proteins associated with dynein during mitosis. If augmin is among these proteins, it suggests an interaction between augmin and dynein. ...
Amino Acid Alphabet
... Also protein structures are often tolerant to amino acid substitutions How did the current amino acid alphabet evolve? The standard alphabet may consist of the smallest and cheapest amino acids that form a functional protein library Helps identify homologous proteins and is crucial for homology mode ...
... Also protein structures are often tolerant to amino acid substitutions How did the current amino acid alphabet evolve? The standard alphabet may consist of the smallest and cheapest amino acids that form a functional protein library Helps identify homologous proteins and is crucial for homology mode ...
A toolbox for validation of mass spectrometry peptides identification
... in the significant category. Dynamic classification has an impact on protein hits: when a PSM is declared ambiguous, it becomes irrelevant to proteins identification. IRMa takes into account these changes to ensure consistency of information such as protein coverage and identification score and dyna ...
... in the significant category. Dynamic classification has an impact on protein hits: when a PSM is declared ambiguous, it becomes irrelevant to proteins identification. IRMa takes into account these changes to ensure consistency of information such as protein coverage and identification score and dyna ...
ImmunO™ Rabbit, Anti-S19 Ribosomal Protein Catalog #: 63659 Lot
... Form:Lyophilized. The antibody (100 ug) is supplied in PBS with 1.0% BSA as a stabilizer and 0.1% Proclin as preservative. Reconstitution:Reconstitute with 100 ul of distilled or de-ionized water. Concentration:1 ug/ul Preparation:Rabbits were immunized with S19 recombinant protein. The antibody was ...
... Form:Lyophilized. The antibody (100 ug) is supplied in PBS with 1.0% BSA as a stabilizer and 0.1% Proclin as preservative. Reconstitution:Reconstitute with 100 ul of distilled or de-ionized water. Concentration:1 ug/ul Preparation:Rabbits were immunized with S19 recombinant protein. The antibody was ...
Aim of the lab - Institute of Microelectronics
... 2.3.2 Non-gel based protein analysis methods (Instructor Spiros D. Garbis) Separation of proteins and peptides with high performance liquid chromatography (HPLC) Combining two or more chromatographic chemistries for peptide/protein separation (multi dimensional HPLC approaches) Introduction of ...
... 2.3.2 Non-gel based protein analysis methods (Instructor Spiros D. Garbis) Separation of proteins and peptides with high performance liquid chromatography (HPLC) Combining two or more chromatographic chemistries for peptide/protein separation (multi dimensional HPLC approaches) Introduction of ...
Chemotaxis pahtway How can physics help?
... Docking (by biochemistry and sequences) • Based on affinity of amino acids. – 1) Define the contacts aminoacids – 2) Produce a score based on the affinity for each sequence. – 3) Score the possible matches – 4) Select the best scores. ...
... Docking (by biochemistry and sequences) • Based on affinity of amino acids. – 1) Define the contacts aminoacids – 2) Produce a score based on the affinity for each sequence. – 3) Score the possible matches – 4) Select the best scores. ...
Structural proteomics of the cell envelope of Gram
... membrane, the periplasmic space, and the outer membrane, can be viewed as a model organelle with a large number of diverse critical functions for bacterial physiology. A significant number of protein structures of both the inner and outer membrane, as well as proteins from the periplasm, have been so ...
... membrane, the periplasmic space, and the outer membrane, can be viewed as a model organelle with a large number of diverse critical functions for bacterial physiology. A significant number of protein structures of both the inner and outer membrane, as well as proteins from the periplasm, have been so ...
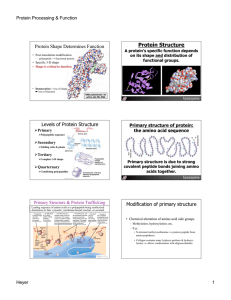
Protein Structure
... Ribonuclease can renature itself. This makes it an unusually tough protein. ...
... Ribonuclease can renature itself. This makes it an unusually tough protein. ...
Protein Expression and Purification Quotation Request Form
... 617 Hutton Street, Suite 101, Raleigh NC 27606-6301 ...
... 617 Hutton Street, Suite 101, Raleigh NC 27606-6301 ...
cell - Zoology, UBC
... Transport vesicles bud off the ER and are transported to the forming face of the Golgi. Membrane-bound proteins and secretory proteins then move through the Golgi, where they are modified, usually by ...
... Transport vesicles bud off the ER and are transported to the forming face of the Golgi. Membrane-bound proteins and secretory proteins then move through the Golgi, where they are modified, usually by ...
Effects of aggregating agents in protein misfolding. An infrared
... Protein misfolding, which include the formation of amyloid aggregates, insoluble aggregates resistant to degradation, are related to a large number of different diseases, mostly neurodegenerative. In this work, hen egg white lysozyme has been used as model because it is a good characterized protein ...
... Protein misfolding, which include the formation of amyloid aggregates, insoluble aggregates resistant to degradation, are related to a large number of different diseases, mostly neurodegenerative. In this work, hen egg white lysozyme has been used as model because it is a good characterized protein ...
Answer Key
... Protein requirements for an athlete trying to increase mass are: a. 0.5 grams/pound body weight b. 0.9-1.0 grams/pound body weight c. 2 grams/pound body weight d. 3 grams/pound body weight ...
... Protein requirements for an athlete trying to increase mass are: a. 0.5 grams/pound body weight b. 0.9-1.0 grams/pound body weight c. 2 grams/pound body weight d. 3 grams/pound body weight ...
This presentation follows on from the talk presented
... In Mascot Insight, we have implemented a report which allows you to use the protein hit sequences from a selected search result to carry out a BLAST like sequence homology search using de novo solutions, in order to try to find additional possible matches to spectra from your dataset, drilling down ...
... In Mascot Insight, we have implemented a report which allows you to use the protein hit sequences from a selected search result to carry out a BLAST like sequence homology search using de novo solutions, in order to try to find additional possible matches to spectra from your dataset, drilling down ...
protein
... _____________________________ _____________________________ _____________________________ _____________________________ It also contains small amounts of: _____________________________ _____________________________ What is the chemical composition of protein? F&N-Protein-Year 9 ...
... _____________________________ _____________________________ _____________________________ _____________________________ It also contains small amounts of: _____________________________ _____________________________ What is the chemical composition of protein? F&N-Protein-Year 9 ...
slides
... contacts in proteins leave an evolutionary record Although evolutionary couplings show promise for the identification of functional sites, homomultimer contacts, alternative conformations and functional sites, many of the predicted contacts involved in these protein features may appear as false posi ...
... contacts in proteins leave an evolutionary record Although evolutionary couplings show promise for the identification of functional sites, homomultimer contacts, alternative conformations and functional sites, many of the predicted contacts involved in these protein features may appear as false posi ...
Protein Requirements for Good Nutrition
... This is where the hard part comes in. You now have two choices. The first is to buy a reputable quality brand of dog food for the activity level of your dog and hope that its needs are being met. This is what most owners do and the average dog does just fine. But if you have a dog with special prote ...
... This is where the hard part comes in. You now have two choices. The first is to buy a reputable quality brand of dog food for the activity level of your dog and hope that its needs are being met. This is what most owners do and the average dog does just fine. But if you have a dog with special prote ...
Protein mass spectrometry

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important emerging method for the characterization of proteins. The two primary methods for ionization of whole proteins are electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). In keeping with the performance and mass range of available mass spectrometers, two approaches are used for characterizing proteins. In the first, intact proteins are ionized by either of the two techniques described above, and then introduced to a mass analyzer. This approach is referred to as ""top-down"" strategy of protein analysis. In the second, proteins are enzymatically digested into smaller peptides using a protease such as trypsin. Subsequently these peptides are introduced into the mass spectrometer and identified by peptide mass fingerprinting or tandem mass spectrometry. Hence, this latter approach (also called ""bottom-up"" proteomics) uses identification at the peptide level to infer the existence of proteins.Whole protein mass analysis is primarily conducted using either time-of-flight (TOF) MS, or Fourier transform ion cyclotron resonance (FT-ICR). These two types of instrument are preferable here because of their wide mass range, and in the case of FT-ICR, its high mass accuracy. Mass analysis of proteolytic peptides is a much more popular method of protein characterization, as cheaper instrument designs can be used for characterization. Additionally, sample preparation is easier once whole proteins have been digested into smaller peptide fragments. The most widely used instrument for peptide mass analysis are the MALDI time-of-flight instruments as they permit the acquisition of peptide mass fingerprints (PMFs) at high pace (1 PMF can be analyzed in approx. 10 sec). Multiple stage quadrupole-time-of-flight and the quadrupole ion trap also find use in this application.