Alpha-COPI Coatomer Protein Is Required for Rough Endoplasmic
... LC-MS/MS analysis and protein annotation (Figure 4). The SDS PAGE analysis revealed that intact proteins of up to ,130 kDa were present in both samples, suggesting that protease activity was minimal in the midgut protein sample from fed mosquitoes. Moreover, no major differences were observed in the ...
... LC-MS/MS analysis and protein annotation (Figure 4). The SDS PAGE analysis revealed that intact proteins of up to ,130 kDa were present in both samples, suggesting that protease activity was minimal in the midgut protein sample from fed mosquitoes. Moreover, no major differences were observed in the ...
Chemistry 501 Lecture 3 Amino Acids
... • Glutathione is -glutamyl-L-cystenylglycine • Two pentapeptides found in the brain are enkaphalins which are natural analgesics – Tyr-Gly-Gly-Phe-Leu (leucine enkaphalin) – Tyr-Gly-Gly-Phe-Met (Methionine enkaphalin) • Opiates bind to the same receptors in the brain intended for the enkaphalins a ...
... • Glutathione is -glutamyl-L-cystenylglycine • Two pentapeptides found in the brain are enkaphalins which are natural analgesics – Tyr-Gly-Gly-Phe-Leu (leucine enkaphalin) – Tyr-Gly-Gly-Phe-Met (Methionine enkaphalin) • Opiates bind to the same receptors in the brain intended for the enkaphalins a ...
Task - Illustrative Mathematics
... This is a full-blown modeling problem (SMP 4). Students must come up with the idea on their own that they can set up two equations in two unknowns to solve the problem, and they must then read, understand and extract information from the nutrition labels to set up the equations in the system (SMP 1) ...
... This is a full-blown modeling problem (SMP 4). Students must come up with the idea on their own that they can set up two equations in two unknowns to solve the problem, and they must then read, understand and extract information from the nutrition labels to set up the equations in the system (SMP 1) ...
Package `PPInfer`
... Description Interactions between proteins occur in many, if not most, biological processes. Most proteins perform their functions in networks associated with other proteins and other biomolecules. This fact has motivated the development of a variety of experimental methods for the identification of ...
... Description Interactions between proteins occur in many, if not most, biological processes. Most proteins perform their functions in networks associated with other proteins and other biomolecules. This fact has motivated the development of a variety of experimental methods for the identification of ...
Document
... operator sites, O64 and OL. The N-terminal domain mediates the interaction with RNA polymerase (lysogenic promoter for repressor maintenance) and thus, activating the CI gene. (Bell et al.) Therefore, above reasons are why the structure of repressor protein (CI, containing CTD and NTD) is important ...
... operator sites, O64 and OL. The N-terminal domain mediates the interaction with RNA polymerase (lysogenic promoter for repressor maintenance) and thus, activating the CI gene. (Bell et al.) Therefore, above reasons are why the structure of repressor protein (CI, containing CTD and NTD) is important ...
Modified Strains PJ69-7A and PJ69-7B for the Yeast Two
... interactions. The technique has been used to establish physical interactions between genetically identified proteins, to identify the components of multiprotein complexes, and to map specific domains within proteins responsible for an interaction. However, there are several limitations to the two-hy ...
... interactions. The technique has been used to establish physical interactions between genetically identified proteins, to identify the components of multiprotein complexes, and to map specific domains within proteins responsible for an interaction. However, there are several limitations to the two-hy ...
Molecular Interaction of PICKI and ATXN3
... deubiquitinating mechanism. ATXN3 was screened as an interactor of PICK1 through yeast-two hybrid, an adaptor protein with important functions in protein trafficking inside the central nervous system and other various organs. PICK1 was found to mediate neurodegeneration of SCA3 as well as other neur ...
... deubiquitinating mechanism. ATXN3 was screened as an interactor of PICK1 through yeast-two hybrid, an adaptor protein with important functions in protein trafficking inside the central nervous system and other various organs. PICK1 was found to mediate neurodegeneration of SCA3 as well as other neur ...
Tumor Necrosis Factor- alpha (human, recombinant) CATALOG NO
... ACTIVITY: Fully biologically active. The ED50, as determined by the cytolysis of murine L929 cells in the presence of Actinomycin D, is less then 0.05 ng/ml, corresponding to a Specific Activity of 2 x 107 IU/mg. SUPPLIED AS: Sterile, filtered white lyophilized powder. The protein was lyophilized af ...
... ACTIVITY: Fully biologically active. The ED50, as determined by the cytolysis of murine L929 cells in the presence of Actinomycin D, is less then 0.05 ng/ml, corresponding to a Specific Activity of 2 x 107 IU/mg. SUPPLIED AS: Sterile, filtered white lyophilized powder. The protein was lyophilized af ...
Slide 1
... Isolate mRNA from NucC+ and NucC- cells Probe a spotted array of Serratia marcescens DNA fragments Characterize clones showing differential regulation ...
... Isolate mRNA from NucC+ and NucC- cells Probe a spotted array of Serratia marcescens DNA fragments Characterize clones showing differential regulation ...
Early states during protein folding - The Astbury Centre for Structural
... To understand how a protein folds, we need to define the properties of the folding landscape and how the polypeptide chain moves through this landscape. For example, does a protein fold quickly on a smooth landscape directly from the unfolded to the folded state, or is the landscape more rugged, inv ...
... To understand how a protein folds, we need to define the properties of the folding landscape and how the polypeptide chain moves through this landscape. For example, does a protein fold quickly on a smooth landscape directly from the unfolded to the folded state, or is the landscape more rugged, inv ...
Leukaemia Section t(11;14)(q23;q32) Atlas of Genetics and Cytogenetics in Oncology and Haematology
... 36 exons, multiple transcripts 13-15 kb. Protein 3969 amino acids; 431 kDa; contains two DNA binding motifs (a AT hook and a CXXC domain), a DNA methyl transferase motif, a bromodomain. MLL is cleaved by taspase 1 into 2 proteins before entering the nucleus, called MLL-N and MLL-C. The FYRN and a FR ...
... 36 exons, multiple transcripts 13-15 kb. Protein 3969 amino acids; 431 kDa; contains two DNA binding motifs (a AT hook and a CXXC domain), a DNA methyl transferase motif, a bromodomain. MLL is cleaved by taspase 1 into 2 proteins before entering the nucleus, called MLL-N and MLL-C. The FYRN and a FR ...
Diseases of the Immune System lec.4
... elevated SAA levels, and ultimately the AA form of amyloid deposits. 3. Aβ amyloid is found in the cerebral lesions of Alzheimer disease. Aβ is a peptide that constitutes the core of cerebral plaques and the amyloid deposits in cerebral blood vessels in this disease. 4. Transthyretin (TTR) is a norm ...
... elevated SAA levels, and ultimately the AA form of amyloid deposits. 3. Aβ amyloid is found in the cerebral lesions of Alzheimer disease. Aβ is a peptide that constitutes the core of cerebral plaques and the amyloid deposits in cerebral blood vessels in this disease. 4. Transthyretin (TTR) is a norm ...
Translation - My Teacher Pages
... • There are 64 possible three-base codons that can be created using the four nitrogen bases (4 x 4 x 4 = 64) • There are 20 different amino acids – There are multiple codons that code for the same amino acids • AUG= start codon for protein synthesis or methionine. This means that translation always ...
... • There are 64 possible three-base codons that can be created using the four nitrogen bases (4 x 4 x 4 = 64) • There are 20 different amino acids – There are multiple codons that code for the same amino acids • AUG= start codon for protein synthesis or methionine. This means that translation always ...
Week 5 - profiles, HMM
... • Hidden Markov models are statistical models that were initially developed for speech recognition. • The most popular use of HMM in molecular biology is as a ‘probabilistic profile’ of a protein family, which is called a profile ...
... • Hidden Markov models are statistical models that were initially developed for speech recognition. • The most popular use of HMM in molecular biology is as a ‘probabilistic profile’ of a protein family, which is called a profile ...
Reprogramming Cells to Fight Disease
... drug, and if Moderna is successful, it could produce tens, hundreds, or even thousands of successful mRNA drugs. How is Moderna able to develop an entire platform, while protein companies could only develop drugs one at a time? Individual proteins are completely different molecules from each other, ...
... drug, and if Moderna is successful, it could produce tens, hundreds, or even thousands of successful mRNA drugs. How is Moderna able to develop an entire platform, while protein companies could only develop drugs one at a time? Individual proteins are completely different molecules from each other, ...
Theory and practice of size exclusion chromatography for
... immunogenicity [12,13]. In recent studies, cytotoxic effects have also been observed with several biotherapeutic proteins due to significant denaturation or chemical alterations of the native protein [8,9,14]. In this context, the characterization of protein modifications requires a number of analytic ...
... immunogenicity [12,13]. In recent studies, cytotoxic effects have also been observed with several biotherapeutic proteins due to significant denaturation or chemical alterations of the native protein [8,9,14]. In this context, the characterization of protein modifications requires a number of analytic ...
Sequencing Grade Modified Trypsin, Frozen, Product Information
... Description: Trypsin specifically hydrolyzes peptide bonds at the carboxyl side of lysine and arginine residues. Unmodified trypsin is subject to auto-proteolysis, generating fragments that can interfere with protein sequencing or HPLC peptide analysis. In addition, auto-proteolysis can result in th ...
... Description: Trypsin specifically hydrolyzes peptide bonds at the carboxyl side of lysine and arginine residues. Unmodified trypsin is subject to auto-proteolysis, generating fragments that can interfere with protein sequencing or HPLC peptide analysis. In addition, auto-proteolysis can result in th ...
Biosynthesis of Protein or Translation
... The amino acids have no direct affinity for mRNA, so tRNA act as an adapter molecule, which recognizes an amino acid on one end and its corresponding codon on the other, is required for translation. ...
... The amino acids have no direct affinity for mRNA, so tRNA act as an adapter molecule, which recognizes an amino acid on one end and its corresponding codon on the other, is required for translation. ...
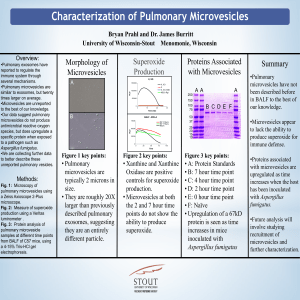
Bryan 2012 Research Day Poster
... Fig. 3: Protein analysis of pulmonary microvesicle samples at different time points from BALF of C57 mice, using a 4-15% Tris-HCl gel electrophoresis. ...
... Fig. 3: Protein analysis of pulmonary microvesicle samples at different time points from BALF of C57 mice, using a 4-15% Tris-HCl gel electrophoresis. ...
Manipulation of Epitope Function by Modification of Peptide
... We have studied the influence of -amino acid substitution in the flanking region on the antibody recognition of the 19TGTQ22 epitope core.6 Analogue peptides corresponding to the optimal epitope sequence (16PTPTGTQ22) have been prepared by the replacement of single or multiple -amino acid residues ...
... We have studied the influence of -amino acid substitution in the flanking region on the antibody recognition of the 19TGTQ22 epitope core.6 Analogue peptides corresponding to the optimal epitope sequence (16PTPTGTQ22) have been prepared by the replacement of single or multiple -amino acid residues ...
Notes for using PROTPOL.f
... (note that the RESTYPE and NORES for the CA, C, O are incorrectly given as those for the following residue – these parameters are only correct for the N) Followed by three lines with (Q(I, I), (H(J, I), J = 1, 3), I = 1, 3) Q (I, I) = Ith diagonal element of moment of inertia matrix in principal axi ...
... (note that the RESTYPE and NORES for the CA, C, O are incorrectly given as those for the following residue – these parameters are only correct for the N) Followed by three lines with (Q(I, I), (H(J, I), J = 1, 3), I = 1, 3) Q (I, I) = Ith diagonal element of moment of inertia matrix in principal axi ...
NUCLEAR PROTEINS II. Similarity of Nonhistone Proteins in
... that in mouse liver nuclei the nuclear sap and n o n h i s t o n e proteins exhibited many similar bands, suggesting that they were not distinct entities. T h e nuclei a p p e a r e d to possess several classes of nonhistone proteins, some of which were more tightly b o u n d to D N A than others; b ...
... that in mouse liver nuclei the nuclear sap and n o n h i s t o n e proteins exhibited many similar bands, suggesting that they were not distinct entities. T h e nuclei a p p e a r e d to possess several classes of nonhistone proteins, some of which were more tightly b o u n d to D N A than others; b ...
user manual for MS Amanda Standalone
... output The fourth parameter is optional and specifies the name of the produced output file. If the parameter is not defined, then the output filename will be the name of the spectrum file or the folder name with suffix “_output”. In this output file, all data are given as tab separated values. A ...
... output The fourth parameter is optional and specifies the name of the produced output file. If the parameter is not defined, then the output filename will be the name of the spectrum file or the folder name with suffix “_output”. In this output file, all data are given as tab separated values. A ...
Protein-Chemistry_Svar-lektionsuppgifter
... C35 does not affect the binding of the substrate, but affects the transistion state almost as much (75 %) as the final product. Thus, C35 is a catalytic group. H48 has a binding function to a certain degree, but above all H48 has a catalytic function. ...
... C35 does not affect the binding of the substrate, but affects the transistion state almost as much (75 %) as the final product. Thus, C35 is a catalytic group. H48 has a binding function to a certain degree, but above all H48 has a catalytic function. ...
Essential Bioinformatics and Biocomputing
... • We would avoid conclusions like this by looking at the similarity scores. This will be done in more detail later in the course, for now it is important to know that the lower the expected value, the better the match. Anything close or greater than 1 should be observed with suspicion. However, some ...
... • We would avoid conclusions like this by looking at the similarity scores. This will be done in more detail later in the course, for now it is important to know that the lower the expected value, the better the match. Anything close or greater than 1 should be observed with suspicion. However, some ...
Protein mass spectrometry

Protein mass spectrometry refers to the application of mass spectrometry to the study of proteins. Mass spectrometry is an important emerging method for the characterization of proteins. The two primary methods for ionization of whole proteins are electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). In keeping with the performance and mass range of available mass spectrometers, two approaches are used for characterizing proteins. In the first, intact proteins are ionized by either of the two techniques described above, and then introduced to a mass analyzer. This approach is referred to as ""top-down"" strategy of protein analysis. In the second, proteins are enzymatically digested into smaller peptides using a protease such as trypsin. Subsequently these peptides are introduced into the mass spectrometer and identified by peptide mass fingerprinting or tandem mass spectrometry. Hence, this latter approach (also called ""bottom-up"" proteomics) uses identification at the peptide level to infer the existence of proteins.Whole protein mass analysis is primarily conducted using either time-of-flight (TOF) MS, or Fourier transform ion cyclotron resonance (FT-ICR). These two types of instrument are preferable here because of their wide mass range, and in the case of FT-ICR, its high mass accuracy. Mass analysis of proteolytic peptides is a much more popular method of protein characterization, as cheaper instrument designs can be used for characterization. Additionally, sample preparation is easier once whole proteins have been digested into smaller peptide fragments. The most widely used instrument for peptide mass analysis are the MALDI time-of-flight instruments as they permit the acquisition of peptide mass fingerprints (PMFs) at high pace (1 PMF can be analyzed in approx. 10 sec). Multiple stage quadrupole-time-of-flight and the quadrupole ion trap also find use in this application.