Scientific poster example - Makerere University News Portal
... entire isolates cluster with previous ASF viruses in genotype IX isolated in Uganda and Kenya using p72 and P54 genes. Analysis of the CVR gene generated three sub-groups one with 23 tetrameric amino acid repeats (TRS) with an additional CAST sequence, the second with 22 TRS while one isolate Ug13. ...
... entire isolates cluster with previous ASF viruses in genotype IX isolated in Uganda and Kenya using p72 and P54 genes. Analysis of the CVR gene generated three sub-groups one with 23 tetrameric amino acid repeats (TRS) with an additional CAST sequence, the second with 22 TRS while one isolate Ug13. ...
Lecture_note_463BI
... -The dbSNP accepts apparently neutral polymorphisms, polymorphisms corresponding to known phenotypes, and regions of no variation. -It was created in September 1998 to supplement GenBank (NCBI’s nucleic acid and protein sequences) Goal Its goal is to act as a single database that contains all identi ...
... -The dbSNP accepts apparently neutral polymorphisms, polymorphisms corresponding to known phenotypes, and regions of no variation. -It was created in September 1998 to supplement GenBank (NCBI’s nucleic acid and protein sequences) Goal Its goal is to act as a single database that contains all identi ...
Powerpoints
... Neural Network – most complex – can detect rules and relationships other methods can’t. Good use cases include those with large number of inputs and relatively few output: Text mining, (Stock) market analysis, manufacturing processes. ...
... Neural Network – most complex – can detect rules and relationships other methods can’t. Good use cases include those with large number of inputs and relatively few output: Text mining, (Stock) market analysis, manufacturing processes. ...
Introduction to the Holonomic Gradient Method in Statistics
... The holonomic gradient method introduced by Nakayama et al. (2011) presents a new methodology for evaluating normalizing constants of probability distributions and for obtaining the maximum likelihood estimate of a statistical model. The method utilizes partial differential equations satisfied by th ...
... The holonomic gradient method introduced by Nakayama et al. (2011) presents a new methodology for evaluating normalizing constants of probability distributions and for obtaining the maximum likelihood estimate of a statistical model. The method utilizes partial differential equations satisfied by th ...
Dot plot - TeachLine
... between different AAs - # of mutations, chemical similarity, PAM matrix ...
... between different AAs - # of mutations, chemical similarity, PAM matrix ...
grappa - Department of Computer Science
... random events (based upon 120 genes and inversion only, but robust to errors in the model) . – Polynomial time, fastest of the three. ...
... random events (based upon 120 genes and inversion only, but robust to errors in the model) . – Polynomial time, fastest of the three. ...
Q 5.2 : Consider the rough guide to worst
... rectangle for rectangle T and L. Add it to the set of rectangles in Fig. 6.29 (pp. 253) and Fig. 6.30 (pp. 254). Redraw the R-tree and R+tree in Fig. 6.29 and 6.30 if this new rectangle is inserted. Briefly justify your solutions by recalling how R-tree and R+tree deal with large objects based on na ...
... rectangle for rectangle T and L. Add it to the set of rectangles in Fig. 6.29 (pp. 253) and Fig. 6.30 (pp. 254). Redraw the R-tree and R+tree in Fig. 6.29 and 6.30 if this new rectangle is inserted. Briefly justify your solutions by recalling how R-tree and R+tree deal with large objects based on na ...
Supporting Online Material for
... 10) In the above tree, assume that the ancestor was an herb (not a tree) without leaves or seeds. Based on the tree and assuming that all evolutionary changes in these traits are shown, which of the tips has a tree habit and lacks true leaves? a) Lepidodendron b) Clubmoss c) Oak d) Psilotum e) Fern ...
... 10) In the above tree, assume that the ancestor was an herb (not a tree) without leaves or seeds. Based on the tree and assuming that all evolutionary changes in these traits are shown, which of the tips has a tree habit and lacks true leaves? a) Lepidodendron b) Clubmoss c) Oak d) Psilotum e) Fern ...
Applying Analytics to Search Engine Marketing (SEM)
... Easy to follow path from root node to a leaf – explanation for any prediction is easy to understand Trees carve up & cover completely the multi-dimensional space – enabling us to assign any new record to an outcome based on which region it falls into. Robust/insensitive to outliers, missing va ...
... Easy to follow path from root node to a leaf – explanation for any prediction is easy to understand Trees carve up & cover completely the multi-dimensional space – enabling us to assign any new record to an outcome based on which region it falls into. Robust/insensitive to outliers, missing va ...
Challenge Lesson Analyzing DNA
... This brings you to a database called the NCBI Entrez database, which contains the sequence of every gene that has ever been sequenced. 2. In the search window at the top of the page, first select the word “Nucleotide” from the pull-down bar. Then, in the text box next to the pull-down bar, type in “ ...
... This brings you to a database called the NCBI Entrez database, which contains the sequence of every gene that has ever been sequenced. 2. In the search window at the top of the page, first select the word “Nucleotide” from the pull-down bar. Then, in the text box next to the pull-down bar, type in “ ...
1-π`1
... and get a topic there. • Otherwise it’s very much like DP sampling. • Since is discrete and we are likely to draw the same component again, they are shared across the groups. • Sharing will not happen if was continuous. ...
... and get a topic there. • Otherwise it’s very much like DP sampling. • Since is discrete and we are likely to draw the same component again, they are shared across the groups. • Sharing will not happen if was continuous. ...
Introduction to Bioinformatics
... The purpose of this exercise is to introduce to you and familiarize you with the National Center for Biotechnology Information (NCBI) web sites. These web sites contain nucleotide and protein databases containing a vast amount of sequencing data that is increasing every single day. As more and more ...
... The purpose of this exercise is to introduce to you and familiarize you with the National Center for Biotechnology Information (NCBI) web sites. These web sites contain nucleotide and protein databases containing a vast amount of sequencing data that is increasing every single day. As more and more ...
Tree Structure for Set of Intervals
... Root nor any node on the left or right boundary path of the tree can have any intervals of the canonical interval decomposition attached to it because their node intervals are unbounded and we are representing only finite intervals. ...
... Root nor any node on the left or right boundary path of the tree can have any intervals of the canonical interval decomposition attached to it because their node intervals are unbounded and we are representing only finite intervals. ...
wg: Use primers wg550F and wgABRZ with cycler profile ST
... specimens. Near references were chosen as species closely related to, but likely distinct from, the museum specimens, based upon the work by [6,7]. Reference sequences came from the same two studies. The near references used were Bembidion paraenulum Maddison (specimen 1856) for B. subfusum; Bembidi ...
... specimens. Near references were chosen as species closely related to, but likely distinct from, the museum specimens, based upon the work by [6,7]. Reference sequences came from the same two studies. The near references used were Bembidion paraenulum Maddison (specimen 1856) for B. subfusum; Bembidi ...
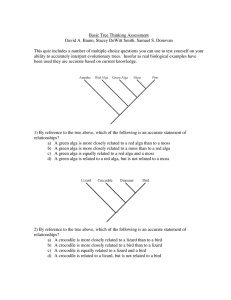
Basic Tree Thinking Assessment David A. Baum, Stacey DeWitt
... 10) In the above tree, assume that the ancestor was an herb (not a tree) without leaves or seeds. Based on the tree and assuming that all evolutionary changes in these traits are shown, which of the tips has a tree habit and lacks true leaves? a) Lepidodendron b) Clubmoss c) Oak d) Psilotum e) Fern ...
... 10) In the above tree, assume that the ancestor was an herb (not a tree) without leaves or seeds. Based on the tree and assuming that all evolutionary changes in these traits are shown, which of the tips has a tree habit and lacks true leaves? a) Lepidodendron b) Clubmoss c) Oak d) Psilotum e) Fern ...