Site-selective incorporation and ligation of
... subsequent hydrolysis of a cysteine residue that is located within a consensus sequence of CxPxR (where x = any amino acid). Conversion of the thiol to the aldehyde is carried out by the Formylglycine Generating Enzyme (FGE). In nature, this transformation is essential for the enzymatic turnover in ...
... subsequent hydrolysis of a cysteine residue that is located within a consensus sequence of CxPxR (where x = any amino acid). Conversion of the thiol to the aldehyde is carried out by the Formylglycine Generating Enzyme (FGE). In nature, this transformation is essential for the enzymatic turnover in ...
Full-Text PDF
... of LUCA (Last Universal Cellular Ancestor), along with the universal conservation of the translation machinery [29]. However, LUCA certainly was not the first life form and most likely, not even the first cellular organism, only an evolutionary bottleneck. The early stages of cellular and especially ...
... of LUCA (Last Universal Cellular Ancestor), along with the universal conservation of the translation machinery [29]. However, LUCA certainly was not the first life form and most likely, not even the first cellular organism, only an evolutionary bottleneck. The early stages of cellular and especially ...
fulltext - DiVA Portal
... within membranes. Other membrane proteins are key players during organelle biogenesis and intracellular trafficking. The protein content in membranes is therefore usually high, being about equal the mass of the lipids6. It is estimated that the human cell contains about 5500 different types of membr ...
... within membranes. Other membrane proteins are key players during organelle biogenesis and intracellular trafficking. The protein content in membranes is therefore usually high, being about equal the mass of the lipids6. It is estimated that the human cell contains about 5500 different types of membr ...
The paradox of elongation factor 4: highly conserved, yet of no
... of E-tRNA: upon back-translocation, the P site will be re-occupied by deacylated tRNA from the E site and 30S subunit P site is therefore maintained by a tRNA, which is a prerequisite for the ribosome to maintain the correct reading frame in all intermediate states of translocation [6]. EF4•GTP bind ...
... of E-tRNA: upon back-translocation, the P site will be re-occupied by deacylated tRNA from the E site and 30S subunit P site is therefore maintained by a tRNA, which is a prerequisite for the ribosome to maintain the correct reading frame in all intermediate states of translocation [6]. EF4•GTP bind ...
Chapter 5 The Structure and Function of Large Biological Molecules
... A) bonding together of several polypeptide chains by weak bonds. B) order in which amino acids are joined in a polypeptide chain. C) unique three-dimensional shape of the fully folded polypeptide. D) organization of a polypeptide chain into an α helix or β pleated sheet. E) overall protein structure ...
... A) bonding together of several polypeptide chains by weak bonds. B) order in which amino acids are joined in a polypeptide chain. C) unique three-dimensional shape of the fully folded polypeptide. D) organization of a polypeptide chain into an α helix or β pleated sheet. E) overall protein structure ...
Deep mutational scanning reveals tail anchor
... Independently of the primary amino acid sequence, the specific codons used to direct ...
... Independently of the primary amino acid sequence, the specific codons used to direct ...
Chemical characterization and in situ nutrient degradability of wet
... Stillage is the byproduct of the distilling industry in which cereal grains are fermented into alcohol (Lee et al., 1991). Traditionally, whole stillage is dried and marketed as distillers' grains with or without solubles. Distillers' grains are identi®ed by the type of grain used in the distillatio ...
... Stillage is the byproduct of the distilling industry in which cereal grains are fermented into alcohol (Lee et al., 1991). Traditionally, whole stillage is dried and marketed as distillers' grains with or without solubles. Distillers' grains are identi®ed by the type of grain used in the distillatio ...
Abstract Background The present study aimed to compare the
... ginseng are 2.4- and 1.9-fold higher compared with cultivated ginseng. Thus, there are notable differences between wild ginseng and cultivated ginseng at the amino acid level. However, the mechanism of these differences and their effect on the medical functions of ginseng are not well understood. Pr ...
... ginseng are 2.4- and 1.9-fold higher compared with cultivated ginseng. Thus, there are notable differences between wild ginseng and cultivated ginseng at the amino acid level. However, the mechanism of these differences and their effect on the medical functions of ginseng are not well understood. Pr ...
Levy APS - Indiana University Bloomington
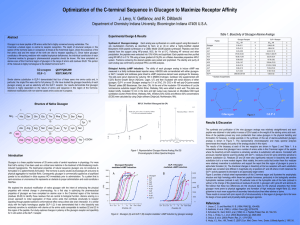
... where the bioactivity at both receptors is extremely sensitive to substitution with alanine. Consistent with previously reported alanine scanning studies with GLP-1, residues 22 and 23 were extremely sensitive to alanine substitution (2). Residues 25 and 26 were also significantly reduced in bioacti ...
... where the bioactivity at both receptors is extremely sensitive to substitution with alanine. Consistent with previously reported alanine scanning studies with GLP-1, residues 22 and 23 were extremely sensitive to alanine substitution (2). Residues 25 and 26 were also significantly reduced in bioacti ...
Attachment 2 - Food Standards Australia New Zealand
... sequence. Polymerase chain reaction (PCR) analyses were performed to verify the sequences at the 5' and 3' ends of the insert. Insert and copy number Southern hybridisation was used to determine the number and nature of DNA insertions in line H7-1. The genomic DNA was digested and probed with an int ...
... sequence. Polymerase chain reaction (PCR) analyses were performed to verify the sequences at the 5' and 3' ends of the insert. Insert and copy number Southern hybridisation was used to determine the number and nature of DNA insertions in line H7-1. The genomic DNA was digested and probed with an int ...
Yeast
... whereas most of accompanying proteins precipitated. It is thus possible that the reduction of almost 35·5% of the total enzyme units found in the crude extract could correspond to Mn SOD, which precipitates with contaminant proteins. When fraction Sf was run on an IMAC separation system (Figure 1), ...
... whereas most of accompanying proteins precipitated. It is thus possible that the reduction of almost 35·5% of the total enzyme units found in the crude extract could correspond to Mn SOD, which precipitates with contaminant proteins. When fraction Sf was run on an IMAC separation system (Figure 1), ...
Get PDF - Wiley Online Library
... Fig. 3. Localization of the PABP-binding site in human eIF4GI. (A) N-terminal deletions. HeLa cells infected with vTF7-3 were co-transfected with pcDNA3-PABP-HA and pcDNA3-GST-eIF4GI fragments indicated in each lane (lanes 1–5) or pcDNA3-GST-CAT (lane 6). One-tenth of the cell extract was used for W ...
... Fig. 3. Localization of the PABP-binding site in human eIF4GI. (A) N-terminal deletions. HeLa cells infected with vTF7-3 were co-transfected with pcDNA3-PABP-HA and pcDNA3-GST-eIF4GI fragments indicated in each lane (lanes 1–5) or pcDNA3-GST-CAT (lane 6). One-tenth of the cell extract was used for W ...
Amino Acid Catabolism: N
... Chime Exercise Two neighboring students or student groups should team up, each displaying one of the following: Transaminase with PLP in Schiff base linkage to the active site lysine residue. Transaminase in the PMP form, with glutarate, an analog of a-ketoglutarate, at the active site. ...
... Chime Exercise Two neighboring students or student groups should team up, each displaying one of the following: Transaminase with PLP in Schiff base linkage to the active site lysine residue. Transaminase in the PMP form, with glutarate, an analog of a-ketoglutarate, at the active site. ...
Highly Conserved Region 141–168 of the NS1 Protein Is a New
... Of the 10 anti-NS1 MAbs described above, 2H11 and 3C4 were found to be complex-specific, 4G1 and 4C2 were found to be serotype-specific, and the others (1A5, 3F10, 4E5, 5E2, 4G11, and 5G12) were found to be subcomplex-specific (Table 2). The MAb isotypes were determined using an immunochromatography ...
... Of the 10 anti-NS1 MAbs described above, 2H11 and 3C4 were found to be complex-specific, 4G1 and 4C2 were found to be serotype-specific, and the others (1A5, 3F10, 4E5, 5E2, 4G11, and 5G12) were found to be subcomplex-specific (Table 2). The MAb isotypes were determined using an immunochromatography ...
Diversity of Amyloid Motifs in NLR Signaling in Fungi
... HeLo and a C-terminal prion forming domain (PFD). The C-terminal prion forming domain (HET-s (218–289)) is necessary and sufficient for prion propagation and has been extensively used as a model system for studying the structure-function relation in amyloid formation and prion propagation. HET-s PFD ...
... HeLo and a C-terminal prion forming domain (PFD). The C-terminal prion forming domain (HET-s (218–289)) is necessary and sufficient for prion propagation and has been extensively used as a model system for studying the structure-function relation in amyloid formation and prion propagation. HET-s PFD ...
EP 1790660 B1
... (a) at least one domain in the protein is deleted and, optionally, (b) no fusion partner is used. [0020] The method will typically involve the steps of: obtaining nucleic acid encoding a protein of the invention; manipulating said nucleic acid to remove at least one domain from within the protein. T ...
... (a) at least one domain in the protein is deleted and, optionally, (b) no fusion partner is used. [0020] The method will typically involve the steps of: obtaining nucleic acid encoding a protein of the invention; manipulating said nucleic acid to remove at least one domain from within the protein. T ...
Are You suprised ?
... is determined by the sequence of nucleotides in a gene. A change in the DNA nucleotide sequence (mutation) of a gene that codes for a protein may result in a change in the amino-acid sequence of the protein. Biochemical evidence of evolution compares favorably with structural evidence of evolution. ...
... is determined by the sequence of nucleotides in a gene. A change in the DNA nucleotide sequence (mutation) of a gene that codes for a protein may result in a change in the amino-acid sequence of the protein. Biochemical evidence of evolution compares favorably with structural evidence of evolution. ...
The 2009 Nobel Prize in Chemistry and KEK`s Photon Factory
... 21 different proteins. The corresponding large subunit, called 50s, consists of two strands of rRNA and up to 34 proteins. These two subunits bind together to form a ribosome called 70S. While the small subunit decodes the genetic information in the mRNA, the large subunit creates a chain of amino a ...
... 21 different proteins. The corresponding large subunit, called 50s, consists of two strands of rRNA and up to 34 proteins. These two subunits bind together to form a ribosome called 70S. While the small subunit decodes the genetic information in the mRNA, the large subunit creates a chain of amino a ...
Optimization of the heterologous expression of folate metabolic Plasmodium falciparum
... E. coli (codon bias). The expression levels of purified hGTPCHI were a greater in comparison to oGTPCHI using the different expression conditions. This is because codon-harmonization involves substituting codons to replicate the codon frequency preference of the target gene in P. falciparum, as suc ...
... E. coli (codon bias). The expression levels of purified hGTPCHI were a greater in comparison to oGTPCHI using the different expression conditions. This is because codon-harmonization involves substituting codons to replicate the codon frequency preference of the target gene in P. falciparum, as suc ...
Bioinformatics in Drug Design
... • Step 1. Dayhoff used 71 protein families, made hypothetical phylogenetic trees and recorded the number of observed substitutions (along each branch of the tree) in a 20x20 target matrix. ...
... • Step 1. Dayhoff used 71 protein families, made hypothetical phylogenetic trees and recorded the number of observed substitutions (along each branch of the tree) in a 20x20 target matrix. ...
Protein structure prediction

Protein structure prediction is the prediction of the three-dimensional structure of a protein from its amino acid sequence — that is, the prediction of its folding and its secondary, tertiary, and quaternary structure from its primary structure. Structure prediction is fundamentally different from the inverse problem of protein design. Protein structure prediction is one of the most important goals pursued by bioinformatics and theoretical chemistry; it is highly important in medicine (for example, in drug design) and biotechnology (for example, in the design of novel enzymes). Every two years, the performance of current methods is assessed in the CASP experiment (Critical Assessment of Techniques for Protein Structure Prediction). A continuous evaluation of protein structure prediction web servers is performed by the community project CAMEO3D.