Name:
... 7. Our next step is translation. What happens during translation and where does this occur? 8. Match the amino acid anticodons with the mRNA codons. (Mr. Mason will ...
... 7. Our next step is translation. What happens during translation and where does this occur? 8. Match the amino acid anticodons with the mRNA codons. (Mr. Mason will ...
Isolation of DNA from 96 Well Plates
... 5. Spin down plate for 3-5 minutes at 3200 rpm to collect condensation (optional). 6. Add 100 μl/well NaCl/Ethanol (@ -20oC). The salt precipitates, so keep the mixture well mixed. Incubate at -20oC for at least 30 minutes until precipitated DNA is visible as long threads under tissue culture micros ...
... 5. Spin down plate for 3-5 minutes at 3200 rpm to collect condensation (optional). 6. Add 100 μl/well NaCl/Ethanol (@ -20oC). The salt precipitates, so keep the mixture well mixed. Incubate at -20oC for at least 30 minutes until precipitated DNA is visible as long threads under tissue culture micros ...
Genetics - California Science Teacher
... 16. Process in which naked DNA is taken up by bacterial or yeast cell 17. Process in which RNA is produced by using a DNA template. 18. Process that results in the production of cDNA from an RNA molecule. 19. Process in which DNA is produced by using a DNA template ...
... 16. Process in which naked DNA is taken up by bacterial or yeast cell 17. Process in which RNA is produced by using a DNA template. 18. Process that results in the production of cDNA from an RNA molecule. 19. Process in which DNA is produced by using a DNA template ...
Molecular markers
... polyacrylamide gels ("sequence gels"); The visualization of the DNA fingerprints by means of autoradiography, phosphoimaging, or other methods. ...
... polyacrylamide gels ("sequence gels"); The visualization of the DNA fingerprints by means of autoradiography, phosphoimaging, or other methods. ...
Protocol for T4 Polynucleotide Kinase, Cloned
... T4 Polynucleotide Kinase (T4 PNK) catalyzes the transfer of the γ-phosphate of ATP to the 5′ terminus of single- and double-stranded DNA or RNA molecules that have a 5′ hydroxyl. The enzyme also removes the 3′ phosphate from 3′-phosphoryl polynucleotides, deoxyribonucleoside 3′-monophosphates, and d ...
... T4 Polynucleotide Kinase (T4 PNK) catalyzes the transfer of the γ-phosphate of ATP to the 5′ terminus of single- and double-stranded DNA or RNA molecules that have a 5′ hydroxyl. The enzyme also removes the 3′ phosphate from 3′-phosphoryl polynucleotides, deoxyribonucleoside 3′-monophosphates, and d ...
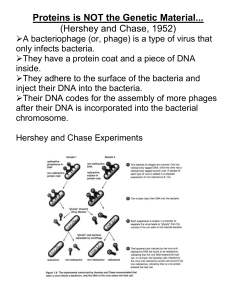
Hershey and Chase`s Experiment
... They adhere to the surface of the bacteria and inject their DNA into the bacteria. Their DNA codes for the assembly of more phages after their DNA is incorporated into the bacterial chromosome. Hershey and Chase Experiments ...
... They adhere to the surface of the bacteria and inject their DNA into the bacteria. Their DNA codes for the assembly of more phages after their DNA is incorporated into the bacterial chromosome. Hershey and Chase Experiments ...
DNA Unit Test Corrections
... 1. In the picture above, what is Arrow #1 pointing to?_________________________ 2. In the picture above, what is Arrow #2 pointing to?_________________________ 3. In the picture above, what is Arrow #3 pointing to?_________________________ 4. Circle a nucleotide. 5. What is the shape of DNA?________ ...
... 1. In the picture above, what is Arrow #1 pointing to?_________________________ 2. In the picture above, what is Arrow #2 pointing to?_________________________ 3. In the picture above, what is Arrow #3 pointing to?_________________________ 4. Circle a nucleotide. 5. What is the shape of DNA?________ ...
downloadable file
... To start, you need a piece of DNA which you want to sequence. Next, you add a DNA priming sequence, the four nucleotides and an enzyme called DNA polymerase which incorporates new nucleotide bases making a new piece of DNA which is a copy of the original piece. In Sanger’s original method, four diff ...
... To start, you need a piece of DNA which you want to sequence. Next, you add a DNA priming sequence, the four nucleotides and an enzyme called DNA polymerase which incorporates new nucleotide bases making a new piece of DNA which is a copy of the original piece. In Sanger’s original method, four diff ...
genome433
... D. SNPs (Single Nucleotide Polymorphisms). A single nucleotide difference between the sequences of two homologous chromosomes (for example, the homologous chromosome 1 copies that you received, one from your mother and one from your father). Most human haploid genomes differ by about 1-3 million ...
... D. SNPs (Single Nucleotide Polymorphisms). A single nucleotide difference between the sequences of two homologous chromosomes (for example, the homologous chromosome 1 copies that you received, one from your mother and one from your father). Most human haploid genomes differ by about 1-3 million ...
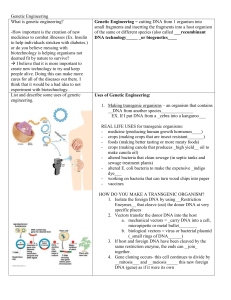
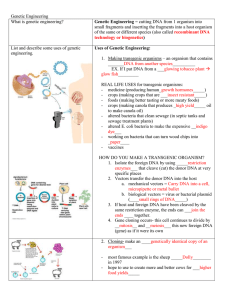
Genetic Engineering Guied Notes
... deemed fit by nature to survive? I believe that it is more important to create new technology to try and keep people alive. Doing this can make more cures for all of the diseases out there. I think that it would be a bad idea to not experiment with biotechnology. List and describe some uses of gen ...
... deemed fit by nature to survive? I believe that it is more important to create new technology to try and keep people alive. Doing this can make more cures for all of the diseases out there. I think that it would be a bad idea to not experiment with biotechnology. List and describe some uses of gen ...
Prepractical demo_SF_Class_2009
... Use another tube for negative control (no DNA) Add primers,nucleotides, Taq DNA polymerase, buffer ...
... Use another tube for negative control (no DNA) Add primers,nucleotides, Taq DNA polymerase, buffer ...
DNA REPLICATION HANDOUT
... 2) Replication Fork: Y-shaped region where new strands of DNA are elongated 3) Okazaki Fragments: Only found on the lagging strand. Since DNA is connected by base pairs, as the original strand “unzips” one of the templates is running in the 5’ to 3’ direction, while the other is 3’ to 5’. As you kno ...
... 2) Replication Fork: Y-shaped region where new strands of DNA are elongated 3) Okazaki Fragments: Only found on the lagging strand. Since DNA is connected by base pairs, as the original strand “unzips” one of the templates is running in the 5’ to 3’ direction, while the other is 3’ to 5’. As you kno ...
NAME CH11 In class assignment Due 2/18/14 Across 1. Initials of
... 3. Separates DNA into fragments by using an electrical current through a gel- ELECTROPHORESIS 4. Circular DNA commonly inserted into bacteria to allow for multiplication- PLASMID 6. Number of loci that the FBI needs from a suspect's DNA- THIRTEEN 10. Organism that contains DNA from a different speci ...
... 3. Separates DNA into fragments by using an electrical current through a gel- ELECTROPHORESIS 4. Circular DNA commonly inserted into bacteria to allow for multiplication- PLASMID 6. Number of loci that the FBI needs from a suspect's DNA- THIRTEEN 10. Organism that contains DNA from a different speci ...
Chapter
... Email: [email protected], Tel.:+84-710 3832475, Fax: +84-7103 831270 Veterinary staff, People’s Committee of Phuoc Hao Commune, Chau Thanh district, Tra Vinh province Postal address: People’s Committee of Phuoc Hao Commune, Chau Thanh district, Tra Vinh province, Vietnam Email: [email protected], ...
... Email: [email protected], Tel.:+84-710 3832475, Fax: +84-7103 831270 Veterinary staff, People’s Committee of Phuoc Hao Commune, Chau Thanh district, Tra Vinh province Postal address: People’s Committee of Phuoc Hao Commune, Chau Thanh district, Tra Vinh province, Vietnam Email: [email protected], ...
EDVOTEK 225 DNA Fingerprinting
... • Used to analyze DNA fragments • DNA has negative charge. • Gel is a sieve which separate DNA fragments according to size, charge and shape. • Only size of DNA affects mobility. • Cleavage of large complex human DNA generates fragments which may exceed resolving capacity of gel. • Cleaved DNA will ...
... • Used to analyze DNA fragments • DNA has negative charge. • Gel is a sieve which separate DNA fragments according to size, charge and shape. • Only size of DNA affects mobility. • Cleavage of large complex human DNA generates fragments which may exceed resolving capacity of gel. • Cleaved DNA will ...
Genetic Engineering
... 1. Isolate the foreign DNA by using _____restriction enzymes___ that cleave (cut) the donor DNA at very specific places 2. Vectors transfer the donor DNA into the host a. mechanical vectors = Carry DNA into a cell, micropipette or metal bullet b. biological vectors = virus or bacterial plasmid (____ ...
... 1. Isolate the foreign DNA by using _____restriction enzymes___ that cleave (cut) the donor DNA at very specific places 2. Vectors transfer the donor DNA into the host a. mechanical vectors = Carry DNA into a cell, micropipette or metal bullet b. biological vectors = virus or bacterial plasmid (____ ...
SEG exam 2 1
... d. will bind to the TATA sequence at the 3’ end of the gene to be transcribed. ____To stimulate translation, the ribosome: a. must bind to the 5’ end of the RNA to be translated. b. must be in the cytoplasm of the cell c. must bind to the 3’ end of the RNA to be translated d. will bind to the TATA s ...
... d. will bind to the TATA sequence at the 3’ end of the gene to be transcribed. ____To stimulate translation, the ribosome: a. must bind to the 5’ end of the RNA to be translated. b. must be in the cytoplasm of the cell c. must bind to the 3’ end of the RNA to be translated d. will bind to the TATA s ...
LECTURE 16 – Using Genomic Variation for Identity DNA Level
... Ø Bacteria can be infected by viruses known as bacteriophage Ø Inbuilt immunity in the bacteria against bacteriophage is the restriction enzyme – cleaves the DNA of the bacteriophage Ø Bacteria protects its own DNA from the restriction enzymes (by methylation) Ø Restriction enzymes cut the DNA leavi ...
... Ø Bacteria can be infected by viruses known as bacteriophage Ø Inbuilt immunity in the bacteria against bacteriophage is the restriction enzyme – cleaves the DNA of the bacteriophage Ø Bacteria protects its own DNA from the restriction enzymes (by methylation) Ø Restriction enzymes cut the DNA leavi ...
Worksheet for Biology 1107 Biological Molecules: Structure and
... 8. What are the monomers of proteins? ...
... 8. What are the monomers of proteins? ...
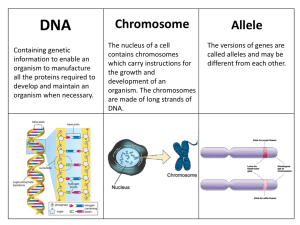
Structure of DNA
... Selectivity of Primers • Primers bind to their complementary sequence on the target DNA – A primer composed of only 3 letter, ACC, for example, would be very likely to encounter its complement in a genome. – As the size of the primer is increased, the likelihood of, for example, a primer sequence o ...
... Selectivity of Primers • Primers bind to their complementary sequence on the target DNA – A primer composed of only 3 letter, ACC, for example, would be very likely to encounter its complement in a genome. – As the size of the primer is increased, the likelihood of, for example, a primer sequence o ...
Honors Biology Final Exam-‐Part 2-‐Semester 2
... 35. The type of inheritance when both versions of the trait are expressed in the heterozygote. 36. Changes in allele frequencies within a population are referred to as: 37. Organisms that look ...
... 35. The type of inheritance when both versions of the trait are expressed in the heterozygote. 36. Changes in allele frequencies within a population are referred to as: 37. Organisms that look ...
5 POINT QUESTIONS 1. A. Give the anticodon sequences (with 5` 3
... C. You isolate the double-stranded DNA genome of a different, unrelated bacterial virus that is also 50 kb in length. When you digest this DNA with BssHI you several times more restriction fragments, with much smaller sizes, than you get from Lambda DNA. Propose a simple explanation for why 2 genomi ...
... C. You isolate the double-stranded DNA genome of a different, unrelated bacterial virus that is also 50 kb in length. When you digest this DNA with BssHI you several times more restriction fragments, with much smaller sizes, than you get from Lambda DNA. Propose a simple explanation for why 2 genomi ...
SNP genotyping

SNP genotyping is the measurement of genetic variations of single nucleotide polymorphisms (SNPs) between members of a species. It is a form of genotyping, which is the measurement of more general genetic variation. SNPs are one of the most common types of genetic variation. An SNP is a single base pair mutation at a specific locus, usually consisting of two alleles (where the rare allele frequency is >1%). SNPs are found to be involved in the etiology of many human diseases and are becoming of particular interest in pharmacogenetics. Because SNPs are conserved during evolution, they have been proposed as markers for use in quantitative trait loci (QTL) analysis and in association studies in place of microsatellites. The use of SNPs is being extended in the HapMap project, which aims to provide the minimal set of SNPs needed to genotype the human genome. SNPs can also provide a genetic fingerprint for use in identity testing. The increase in interest in SNPs has been reflected by the furious development of a diverse range of SNP genotyping methods.