No Slide Title
... normal B cells, but many other tested genes are not mutated at higher rates. •Many lymphoid tumors involve breakpoints between Ig genes and oncogenes. Some of these are associated with V(D)J type recombination, others with class switch, others with functional V regions. ...
... normal B cells, but many other tested genes are not mutated at higher rates. •Many lymphoid tumors involve breakpoints between Ig genes and oncogenes. Some of these are associated with V(D)J type recombination, others with class switch, others with functional V regions. ...
Analytical challenges in the genetic diagnosis of Lynch
... the same effect at the protein level, i.e., exon 5 skipping (p.Val265_Gln314del). Consequently, important functional domains of the protein were affected, such as MutS II and III, which are connector and lever domains, respectively. These domains play different roles in holding the DNA that is to be ...
... the same effect at the protein level, i.e., exon 5 skipping (p.Val265_Gln314del). Consequently, important functional domains of the protein were affected, such as MutS II and III, which are connector and lever domains, respectively. These domains play different roles in holding the DNA that is to be ...
7 Self study questions
... 1. Explain why ORF scanning is a feasible way of identifying genes in a prokaryotic DNA sequence. 2. What modifications are introduced when ORF scanning is applied to a eukaryotic DNA sequence? 3. Describe how homology searching is used to locate genes in a DNA sequence and to assign possible functi ...
... 1. Explain why ORF scanning is a feasible way of identifying genes in a prokaryotic DNA sequence. 2. What modifications are introduced when ORF scanning is applied to a eukaryotic DNA sequence? 3. Describe how homology searching is used to locate genes in a DNA sequence and to assign possible functi ...
Introduction - Milan Area Schools
... • An approach called positional cloning can be used when no candidate protein or deletion is known for a gene. • Reference points for positional cloning are genetic markers on the DNA. • Restriction enzymes are used to cut DNA molecules at specific recognition sequences. • If a sequence is not recog ...
... • An approach called positional cloning can be used when no candidate protein or deletion is known for a gene. • Reference points for positional cloning are genetic markers on the DNA. • Restriction enzymes are used to cut DNA molecules at specific recognition sequences. • If a sequence is not recog ...
Understanding genetic counseling and testing
... Cancer genetic counseling is the process of collecting your detailed personal and family history, assessing your personal risk, and discussing your genetic testing options. These services should be provided by a counselor who is board-certified by the American Board of Genetic Counseling. Genetic te ...
... Cancer genetic counseling is the process of collecting your detailed personal and family history, assessing your personal risk, and discussing your genetic testing options. These services should be provided by a counselor who is board-certified by the American Board of Genetic Counseling. Genetic te ...
Introduction - Cedar Crest College
... In a simple screening test devised by Robert Guthrie in 1963, auxotrophic bacteria are used to detect the presence of phenylalanine. If the test is positive, additional biochemical tests are run. ...
... In a simple screening test devised by Robert Guthrie in 1963, auxotrophic bacteria are used to detect the presence of phenylalanine. If the test is positive, additional biochemical tests are run. ...
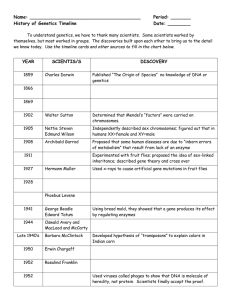
Name
... themselves, but most worked in groups. The discoveries built upon each other to bring us to the detail we know today. Use the timeline cards and other sources to fill in the chart below. ...
... themselves, but most worked in groups. The discoveries built upon each other to bring us to the detail we know today. Use the timeline cards and other sources to fill in the chart below. ...
Unit 4 Mitosis, Meiosis and cell regulation
... Be able to relate what stem cells are and the differences between the types of cloning and the types of stem cells Be able to explain the lac operon and trp operon and their overall goal. Be able to explain the three post-transcriptional modifications of eukaryotic cells. Be able to explain the two ...
... Be able to relate what stem cells are and the differences between the types of cloning and the types of stem cells Be able to explain the lac operon and trp operon and their overall goal. Be able to explain the three post-transcriptional modifications of eukaryotic cells. Be able to explain the two ...
File - Mr. Lambdin`s Biology
... Manipulating DNA • Scientists can cut and paste DNA to get specific sequences that they want • Very similar to cutting and splicing ...
... Manipulating DNA • Scientists can cut and paste DNA to get specific sequences that they want • Very similar to cutting and splicing ...
Artificial Neural Network
... each cancer type • Based on the validation set, for each type of cancer an empirical distribution of its distance is generated • For a given test sample, the system can reject possible classification based on these probability distributions ...
... each cancer type • Based on the validation set, for each type of cancer an empirical distribution of its distance is generated • For a given test sample, the system can reject possible classification based on these probability distributions ...
Gene Section MIR449A (microRNA 449a) Atlas of Genetics and Cytogenetics
... (Noonan et al., 2009; Yang et al., 2009; Lizé et al., 2010; Noonan et al., 2010; Bou Kheir et al., 2011). It is also involved in mucociliary differentiation (Lizé et al., 2010; Marcet et al., 2011). miR-449 regulates several pathways (reviewed in Lizé et al., 2011) including Notch (Capuano et al., 2 ...
... (Noonan et al., 2009; Yang et al., 2009; Lizé et al., 2010; Noonan et al., 2010; Bou Kheir et al., 2011). It is also involved in mucociliary differentiation (Lizé et al., 2010; Marcet et al., 2011). miR-449 regulates several pathways (reviewed in Lizé et al., 2011) including Notch (Capuano et al., 2 ...
Biology 122, Spring 2014 Activities for the week of March 10
... Activities for the week of March 10-14 Directions. In lieu of class on 3/10, 3/12, and 3/14, you should work on the following activities. You should devote the amount of time you spend in class in Biology 122 to these activities, i.e. 150 minutes for the week. You are welcome to work with others on ...
... Activities for the week of March 10-14 Directions. In lieu of class on 3/10, 3/12, and 3/14, you should work on the following activities. You should devote the amount of time you spend in class in Biology 122 to these activities, i.e. 150 minutes for the week. You are welcome to work with others on ...
Name: Date: Period: _____ Unit 6 (DNA, RNA, and Protein
... make an error when pairing new nucleotides with nucleotides on the template strand of DNA. It may match a C with an A, rather than a T with an A. ...
... make an error when pairing new nucleotides with nucleotides on the template strand of DNA. It may match a C with an A, rather than a T with an A. ...
PRESS RELEASE_Novel designed molecules could stop
... CSIC, and from the Novartis Institute for BioMedical Research. "In this project we initially designed inhibitors of the cellular adhesion involved in the metastasis of murine melanoma, and then we carried out the chemical synthesis of these molecules, checking their potency and their biological acti ...
... CSIC, and from the Novartis Institute for BioMedical Research. "In this project we initially designed inhibitors of the cellular adhesion involved in the metastasis of murine melanoma, and then we carried out the chemical synthesis of these molecules, checking their potency and their biological acti ...
An Excel Macro to Visualise Patterns for Chosen Genes
... My Excel Macro Based on macro by Frank Millenaar (Utrecht) Selects genes of interest from Mas5 output spreadsheet Visualises expression profiles as combined: data-tables heat-maps Can adapt for different combinations of chips to give different profiles Eg a range of organs. Similar in function to Ge ...
... My Excel Macro Based on macro by Frank Millenaar (Utrecht) Selects genes of interest from Mas5 output spreadsheet Visualises expression profiles as combined: data-tables heat-maps Can adapt for different combinations of chips to give different profiles Eg a range of organs. Similar in function to Ge ...
NIH Public Access
... problems associated with cytogenetic analysis of epithelial tumors compared to that of hematological cancers [34]. Thus there may be many more epithelial cancer gene fusions that remain to be discovered, either by informatic approaches or by experimental approaches such as high-throughput sequencing ...
... problems associated with cytogenetic analysis of epithelial tumors compared to that of hematological cancers [34]. Thus there may be many more epithelial cancer gene fusions that remain to be discovered, either by informatic approaches or by experimental approaches such as high-throughput sequencing ...
GENETICS SOL REVIEW – 2015 PART II Name ____________________________
... to albino (g). According to the Punnett square, what is the chance of this heterozygous cross producing albino corn plants? A One in four _ B Two in four C Three in four D Four in four ...
... to albino (g). According to the Punnett square, what is the chance of this heterozygous cross producing albino corn plants? A One in four _ B Two in four C Three in four D Four in four ...
16 Mustafa Saffarini NOOR MA`ABREH PATHOLOGY Mazen al
... activated, apoptosis isn’t going to be activated allowing another mutation from passing by that check point and causing damage. This is called a mutator phenotype, where patients who have P53 mutations accumulate further mutations far more easily than patients who have a normal P53 system. ...
... activated, apoptosis isn’t going to be activated allowing another mutation from passing by that check point and causing damage. This is called a mutator phenotype, where patients who have P53 mutations accumulate further mutations far more easily than patients who have a normal P53 system. ...
Oncogenomics
Oncogenomics is a relatively new sub-field of genomics that applies high throughput technologies to characterize genes associated with cancer. Oncogenomics is synonymous with ""cancer genomics"". Cancer is a genetic disease caused by accumulation of mutations to DNA leading to unrestrained cell proliferation and neoplasm formation. The goal of oncogenomics is to identify new oncogenes or tumor suppressor genes that may provide new insights into cancer diagnosis, predicting clinical outcome of cancers, and new targets for cancer therapies. The success of targeted cancer therapies such as Gleevec, Herceptin, and Avastin raised the hope for oncogenomics to elucidate new targets for cancer treatment.Besides understanding the underlying genetic mechanisms that initiates or drives cancer progression, one of the main goals of oncogenomics is to allow for the development of personalized cancer treatment. Cancer develops due to an accumulation of mutations in DNA. These mutations accumulate randomly, and thus, different DNA mutations and mutation combinations exist between different individuals with the same type of cancer. Thus, identifying and targeting specific mutations which have occurred in an individual patient may lead to increased efficacy of cancer therapy.The completion of the Human Genome Project has greatly facilitated the field of oncogenomics and has increased the abilities of researchers to find cancer causing genes. In addition, the sequencing technologies now available for sequence generation and data analysis have been applied to the study of oncogenomics. With the amount of research conducted on cancer genomes and the accumulation of databases documenting the mutational changes, it has been predicted that the most important cancer-causing mutations, rearrangements, and altered expression levels will be cataloged and well characterized within the next decade.Cancer research may look either on the genomic level at DNA mutations, the epigenetic level at methylation or histone modification changes, the transcription level at altered levels of gene expression, or the protein level at altered levels of protein abundance and function in cancer cells. Oncogenomics focuses on the genomic, epigenomic, and transcript level alterations in cancer.