biochem ch 7 [12-11
... function of protein in body o Protein must fold in such a way that it is flexible, stable, able to function in correct site in cell, and capable of being degraded by cellular enzymes Rigidity of peptide backbone determines types of secondary structure that can occur In globular proteins such as ...
... function of protein in body o Protein must fold in such a way that it is flexible, stable, able to function in correct site in cell, and capable of being degraded by cellular enzymes Rigidity of peptide backbone determines types of secondary structure that can occur In globular proteins such as ...
Chapter 31 - Department of Chemistry [FSU]
... polymerase has passed a consensus AAUAAA sequencethe poly(A)+ addition site. • The transcript is cleaved by an endonuclease about 10-30 bases downstream of this consensus site. • Poly(A) polymerase adds 100-200 adenine residues to the ...
... polymerase has passed a consensus AAUAAA sequencethe poly(A)+ addition site. • The transcript is cleaved by an endonuclease about 10-30 bases downstream of this consensus site. • Poly(A) polymerase adds 100-200 adenine residues to the ...
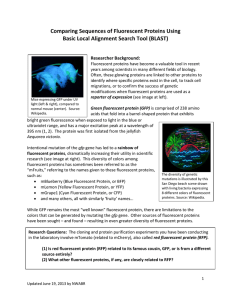
Comparing Sequences of Fluorescent Proteins Using
... have been sought – and found – resulting in even greater diversity of fluorescent proteins. Research Questions: The cloning and protein purification experiments you have been conducting in the laboratory involve mTomato (related to mCherry), also called red fluorescent protein (RFP). (1) Is red fluo ...
... have been sought – and found – resulting in even greater diversity of fluorescent proteins. Research Questions: The cloning and protein purification experiments you have been conducting in the laboratory involve mTomato (related to mCherry), also called red fluorescent protein (RFP). (1) Is red fluo ...
(CH14) Translation (Slides)
... Biochemical evidence that 23S rRNA is a ribozyme • “Fragment reaction” used by Harry Noller and colleagues to shown that purified bacterial 23S rRNA has “peptidyl transferase activity” in vitro. ...
... Biochemical evidence that 23S rRNA is a ribozyme • “Fragment reaction” used by Harry Noller and colleagues to shown that purified bacterial 23S rRNA has “peptidyl transferase activity” in vitro. ...
Proteins are polymers of amino acids, Polypeptides (cofactors
... Proteins are large organic compounds made of amino acids arranged in a linear chain and joined together by peptide bonds between the carboxyl and amino groups of adjacent amino acid residues. They play key roles in constructing and maintaining living cells. The word protein comes from the Gre ...
... Proteins are large organic compounds made of amino acids arranged in a linear chain and joined together by peptide bonds between the carboxyl and amino groups of adjacent amino acid residues. They play key roles in constructing and maintaining living cells. The word protein comes from the Gre ...
CANADIAN TRANSLATION OF FISHERIES AND AQUATIC
... 98 cm, parrs, sexually mature young salmon which have yet to migrate, as well as "early run" salmot in April, sexually inactive adult salmon, smolts, ànd mendeds preparing for the catadromous migration. ...
... 98 cm, parrs, sexually mature young salmon which have yet to migrate, as well as "early run" salmot in April, sexually inactive adult salmon, smolts, ànd mendeds preparing for the catadromous migration. ...
RNA Interference
... Double-stranded RNA can be introduced experimentally to silence target genes interested Nature 418:244-251, 2002 37 ...
... Double-stranded RNA can be introduced experimentally to silence target genes interested Nature 418:244-251, 2002 37 ...
Amino Acid Structure
... called amino acids Every amino acid possesses an amino end and a carboxylic acid end There are twenty different naturally occurring amino acids Amino acids differ by virtue of the nature of their R groups Amino acids bond together forming peptide bonds When two amino acids bond during a co ...
... called amino acids Every amino acid possesses an amino end and a carboxylic acid end There are twenty different naturally occurring amino acids Amino acids differ by virtue of the nature of their R groups Amino acids bond together forming peptide bonds When two amino acids bond during a co ...
Transcription
... within nucleus • snRNA: a class of small RNA molecules within the nucleus snRNA ...
... within nucleus • snRNA: a class of small RNA molecules within the nucleus snRNA ...
2nd Amino Acid Workshop - Maastricht Proteomics Center
... question to be answered but surely also on the available budget because the price of these systems may easily rise above half a million U.S. dollars. In our approach, we prefer to study plasma protein synthesis using the ESI-ion-trap-quad systems. They not only enable quantification of peptide isoto ...
... question to be answered but surely also on the available budget because the price of these systems may easily rise above half a million U.S. dollars. In our approach, we prefer to study plasma protein synthesis using the ESI-ion-trap-quad systems. They not only enable quantification of peptide isoto ...
Directed Evolution of ATP Binding Proteins from a Zinc Finger
... The sequences of the clones discussed above are shown in Figure 3, and the multiple alignment reveals clear sequence patterns. The first random loop of every sequence begins with XVXH, where X is any amino acid. The second random loop of every sequence with the exception of clone 19 begins with XXCX ...
... The sequences of the clones discussed above are shown in Figure 3, and the multiple alignment reveals clear sequence patterns. The first random loop of every sequence begins with XVXH, where X is any amino acid. The second random loop of every sequence with the exception of clone 19 begins with XXCX ...
Protein
... Fats and carbs cannot replace protein Needed to replace wear and tear of tissue and keep up protein concentration in the blood Excess protein, once converted to energy, cannot convert back to protein ...
... Fats and carbs cannot replace protein Needed to replace wear and tear of tissue and keep up protein concentration in the blood Excess protein, once converted to energy, cannot convert back to protein ...
The Bcl-3 oncoprotein acts as a bridging factor between NF
... Speci®c Bcl-3 interactions were con®rmed in each case by reciprocal co-immune precipitations with antisera against Bcl-3 or against the tagged interacting proteins as shown in Figure 1b. When co-translated, an antiBcl-3 antibody co-precipitated Bcl-3 and each of the interacting proteins (lane 5). Si ...
... Speci®c Bcl-3 interactions were con®rmed in each case by reciprocal co-immune precipitations with antisera against Bcl-3 or against the tagged interacting proteins as shown in Figure 1b. When co-translated, an antiBcl-3 antibody co-precipitated Bcl-3 and each of the interacting proteins (lane 5). Si ...
Note
... 2. Polarity and Hydrogen Bonding A. describes the uneven distribution of electrons or uneven electron density of the water molecule hydrogen atom shares one pair of electrons with the oxygen atom and the oxygen atom also has 2 more pairs of electrons that are not shared with any other atom B. the o ...
... 2. Polarity and Hydrogen Bonding A. describes the uneven distribution of electrons or uneven electron density of the water molecule hydrogen atom shares one pair of electrons with the oxygen atom and the oxygen atom also has 2 more pairs of electrons that are not shared with any other atom B. the o ...
1.Lect .AADegradation
... Transaminases are normally intracellular enzymes. They are elevated in the blood when damage to the cells producing these enzymes occurs. * Increase level of both ALT & AST indicates possible damage to the liver cells. * Increase level of AST ALONE suggests damage to heart muscle , skeletal muscle o ...
... Transaminases are normally intracellular enzymes. They are elevated in the blood when damage to the cells producing these enzymes occurs. * Increase level of both ALT & AST indicates possible damage to the liver cells. * Increase level of AST ALONE suggests damage to heart muscle , skeletal muscle o ...
No Slide Title
... The nucleotide bases of the RNA molecule are complementary to the ones on the DNA strand The DNA molecule was used as a template to make mRNA ...
... The nucleotide bases of the RNA molecule are complementary to the ones on the DNA strand The DNA molecule was used as a template to make mRNA ...
Morphologically distinct phenotypes of spermatozoa in infertile men
... OBJECTIVE: Seven morphologically distinct spermatozoal phenotypes can be detected in human semen under electron microscopy: sperm with dysplasia of the fibrous sheath, non-specific flagellar defects, immotile cilia syndrome, acrosomal hypoplasia, defective chromatin condensation and compaction, pin ...
... OBJECTIVE: Seven morphologically distinct spermatozoal phenotypes can be detected in human semen under electron microscopy: sperm with dysplasia of the fibrous sheath, non-specific flagellar defects, immotile cilia syndrome, acrosomal hypoplasia, defective chromatin condensation and compaction, pin ...
Biomolecular chemistry 4. From amino acids to proteins
... • A: Histidine is very good at donating and accepting protons at physiological pH. This is a very important part of many enzyme mechanisms. I may have mentioned that histidine is not such a good nucleophile. For enzyme mechanisms that involve a nucleophilic attack on the substrate, cysteine would be ...
... • A: Histidine is very good at donating and accepting protons at physiological pH. This is a very important part of many enzyme mechanisms. I may have mentioned that histidine is not such a good nucleophile. For enzyme mechanisms that involve a nucleophilic attack on the substrate, cysteine would be ...
interpreted as a demonstration of a biologically significant protein
... A drawback to chemical labeling is that the modified protein may lose one or more of its activities. Hence, the labeled protein must be tested for activity relative to unlabeled protein. Reductive methylation is the least invasive technique because it introduces only one or two methyl groups. In our ...
... A drawback to chemical labeling is that the modified protein may lose one or more of its activities. Hence, the labeled protein must be tested for activity relative to unlabeled protein. Reductive methylation is the least invasive technique because it introduces only one or two methyl groups. In our ...
The trans-Golgi network GRIP-domain proteins form α
... are predicted to adopt a rod-like or extended fibrous structure. Members of the family of coiled-coil proteins specifically located on the Golgi (termed golgins), such as p115 and GM130, have been shown to be important as tethering molecules and in the biogenesis of membranes of the Golgi stack [1–5 ...
... are predicted to adopt a rod-like or extended fibrous structure. Members of the family of coiled-coil proteins specifically located on the Golgi (termed golgins), such as p115 and GM130, have been shown to be important as tethering molecules and in the biogenesis of membranes of the Golgi stack [1–5 ...
Biochemistry 304 2014 Student Edition TRANSCRIPTION
... other small nuclear and cytosolic RNAs, located in nucleoplasm Do not have a σ factor like prokaryotes, but the RNAP is recruited to the initiation site by accessory proteins that recognize promoters. Each RNAP recognizes a different promoter. ...
... other small nuclear and cytosolic RNAs, located in nucleoplasm Do not have a σ factor like prokaryotes, but the RNAP is recruited to the initiation site by accessory proteins that recognize promoters. Each RNAP recognizes a different promoter. ...
Transcription and Translation
... 3. The ribosome moves down the mRNA by one codon, and all three tRNAs move one position within the ribosome. The tRNA in the E site exits; the tRNA in the P site moves to the E site; and the tRNA in the A site switches to the P site. ...
... 3. The ribosome moves down the mRNA by one codon, and all three tRNAs move one position within the ribosome. The tRNA in the E site exits; the tRNA in the P site moves to the E site; and the tRNA in the A site switches to the P site. ...
Defense - Gerstein Lab
... We observe diversity in membrane protein content and abundance, and show that it is a reflection of different environmental conditions more than phylogenetic diversity (16S) These are mostly oligotrophic (nutrient poor) waters and environmental conditions have likely been fairly constant over many y ...
... We observe diversity in membrane protein content and abundance, and show that it is a reflection of different environmental conditions more than phylogenetic diversity (16S) These are mostly oligotrophic (nutrient poor) waters and environmental conditions have likely been fairly constant over many y ...
SR protein

SR proteins are a conserved family of proteins involved in RNA splicing. SR proteins are named because they contain a protein domain with long repeats of serine and arginine amino acid residues, whose standard abbreviations are ""S"" and ""R"" respectively. SR proteins are 50-300 amino acids in length and composed of two domains, the RNA recognition motif (RRM) region and the RS binding domain. SR proteins are more commonly found in the nucleus than the cytoplasm, but several SR proteins are known to shuttle between the nucleus and the cytoplasm.SR proteins were discovered in the 1990s in Drosophila and in amphibian oocytes, and later in humans. In general, metazoans appear to have SR proteins and unicellular organisms lack SR proteins.SR proteins are important in constitutive and alternative pre-mRNA splicing, mRNA export, genome stabilization, nonsense-mediated decay, and translation. SR proteins alternatively splice pre-mRNA by preferentially selecting different splice sites on the pre-mRNA strands to create multiple mRNA transcripts from one pre-mRNA transcript. Once splicing is complete the SR protein may or may not remain attached to help shuttle the mRNA strand out of the nucleus. As RNA Polymerase II is transcribing DNA into RNA, SR proteins attach to newly made pre-mRNA to prevent the pre-mRNA from binding to the coding DNA strand to increase genome stabilization. Topoisomerase I and SR proteins also interact to increase genome stabilization. SR proteins can control the concentrations of specific mRNA that is successfully translated into protein by selecting for nonsense-mediated decay codons during alternative splicing. SR proteins can alternatively splice NMD codons into its own mRNA transcript to auto-regulate the concentration of SR proteins. Through the mTOR pathway and interactions with polyribosomes, SR proteins can increase translation of mRNA.Ataxia telangiectasia, neurofibromatosis type 1, several cancers, HIV-1, and spinal muscular atrophy have all been linked to alternative splicing by SR proteins.
![Chapter 31 - Department of Chemistry [FSU]](http://s1.studyres.com/store/data/017094902_1-c913d8622e17655318ad84818ec4b5cb-300x300.png)