DNA - The Physics Teacher
... distinguish that DNA from other DNA. DNA is extracted from cells e.g. blood or semen by breaking up the cell membrane. DNA amplification can be used if the quantity of DNA is low. Increasing the quantity is done by a technique called the polymerase chain reaction (PCR). Restriction enzymes are ...
... distinguish that DNA from other DNA. DNA is extracted from cells e.g. blood or semen by breaking up the cell membrane. DNA amplification can be used if the quantity of DNA is low. Increasing the quantity is done by a technique called the polymerase chain reaction (PCR). Restriction enzymes are ...
DNA Replication, Recombination, and Repair 2
... are aligned – synapsis. (B) Recombination begins with the introduction of single-stranded nicks at homologous sites on two chromosomes (C) Strand invasion occurs through partial unwinding and basepairing with the intact strand in the other duplex (D) Free ends from different duplexes are ligated res ...
... are aligned – synapsis. (B) Recombination begins with the introduction of single-stranded nicks at homologous sites on two chromosomes (C) Strand invasion occurs through partial unwinding and basepairing with the intact strand in the other duplex (D) Free ends from different duplexes are ligated res ...
Package `microRNA` - USTC Open Source Software Mirror
... A set of human microRNA names and their corresponding known targets given as ensembl Transcript IDs. Usage data(hsTargets) Format A data frame of microRNAs and their target ensembl IDs as recovered from miRBase. Additional columns are also provided to give the Chromosome as well as the start and end ...
... A set of human microRNA names and their corresponding known targets given as ensembl Transcript IDs. Usage data(hsTargets) Format A data frame of microRNAs and their target ensembl IDs as recovered from miRBase. Additional columns are also provided to give the Chromosome as well as the start and end ...
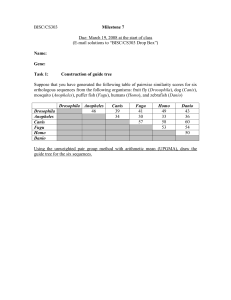
Milestone7
... At the top of the results page, click on the “Start Jalview” button to open an interactive display of the MSA. One of the advantages of a MSA is that it can provide insight into various properties of a family of proteins. When studying your MSA, if you find portions of your sequences that do not ali ...
... At the top of the results page, click on the “Start Jalview” button to open an interactive display of the MSA. One of the advantages of a MSA is that it can provide insight into various properties of a family of proteins. When studying your MSA, if you find portions of your sequences that do not ali ...
Ch. 12 end of chapter review
... DNA polymerase is an enzyme that joins individual nucleotides to produce a new strand of DNA. Replication in most prokaryotic cells starts from a single point and proceeds in two directions until the entire chromosome is copied. In eukaryotic cells, replication may begin at dozens or even hundreds o ...
... DNA polymerase is an enzyme that joins individual nucleotides to produce a new strand of DNA. Replication in most prokaryotic cells starts from a single point and proceeds in two directions until the entire chromosome is copied. In eukaryotic cells, replication may begin at dozens or even hundreds o ...
12.3 How Is Biotechnology Used In Forensic Science?
... • Anyone convicted of certain crimes must give a blood sample. • Forensic technicians then determine the criminal’s DNA profile. • This DNA profile is coded and stored in computer files. • Because all forensic labs use the same STRs, computers can easily determine if DNA left at a crime scene matche ...
... • Anyone convicted of certain crimes must give a blood sample. • Forensic technicians then determine the criminal’s DNA profile. • This DNA profile is coded and stored in computer files. • Because all forensic labs use the same STRs, computers can easily determine if DNA left at a crime scene matche ...
Alignment of amino-acid and nucleotide sequences
... An alignment is an evolutionarily meaningful comparison of two or more sequences (DNA, RNA, or proteins). In the case of two DNA sequences, an alignment consists of a series of paired bases, one base from each sequence. There are three types of pairs: (1) matches = the same nucleotide appears in bo ...
... An alignment is an evolutionarily meaningful comparison of two or more sequences (DNA, RNA, or proteins). In the case of two DNA sequences, an alignment consists of a series of paired bases, one base from each sequence. There are three types of pairs: (1) matches = the same nucleotide appears in bo ...
Molecular analysis of putative genetic factors affecting BSE
... the laboratory. Some of the samples were 6 years old and in very limited supply when used so the purity could not be improved. Given this variability between samples the study was continued by analysing individual samples, rather than pools. This was considerably more labour intensive, but increased ...
... the laboratory. Some of the samples were 6 years old and in very limited supply when used so the purity could not be improved. Given this variability between samples the study was continued by analysing individual samples, rather than pools. This was considerably more labour intensive, but increased ...
Chapter 16 - HCC Learning Web
... Watson and Crick’s semiconservative model of replication predicts that when a double helix replicates, each daughter molecule will have one old strand (derived or “conserved” from the parent molecule) and one newly made strand Competing models were the conservative model (the two parent strands ...
... Watson and Crick’s semiconservative model of replication predicts that when a double helix replicates, each daughter molecule will have one old strand (derived or “conserved” from the parent molecule) and one newly made strand Competing models were the conservative model (the two parent strands ...
(hrM) analysis for mutation screening of genes related to hereditary
... of the region of interest in the presence of a fluorescent dsDNA-binding dye. The dye exhibits high fluorescence when bound to dsDNA and low fluorescence in the unbound state. Following PCR, the product is gradually melted using instrumentation capable of capturing a large number of fluorescent data ...
... of the region of interest in the presence of a fluorescent dsDNA-binding dye. The dye exhibits high fluorescence when bound to dsDNA and low fluorescence in the unbound state. Following PCR, the product is gradually melted using instrumentation capable of capturing a large number of fluorescent data ...
Sample Chapter
... location (in base pairs) and distance between genes or markers, or unknown DNA or genes. These maps provide information about the physical organization of the DNA; examples are the location of restriction enzyme sites and the order of restriction fragments of chromosomes. An entire genome can be stu ...
... location (in base pairs) and distance between genes or markers, or unknown DNA or genes. These maps provide information about the physical organization of the DNA; examples are the location of restriction enzyme sites and the order of restriction fragments of chromosomes. An entire genome can be stu ...
MOLECULAR GENETICS
... about the bases A-t ans C-G. • Wilkins allowed Crick to get Franklin’s information from her lab’s annual report without asking Franklin. He gave Franklin’s X ray pictures to Crick. • They learned all the positions and measurements from Franklin’s picture and build a model in which everything fit exc ...
... about the bases A-t ans C-G. • Wilkins allowed Crick to get Franklin’s information from her lab’s annual report without asking Franklin. He gave Franklin’s X ray pictures to Crick. • They learned all the positions and measurements from Franklin’s picture and build a model in which everything fit exc ...
Enrichment of genes and location of mutations in cloned DNA
... to the isolation of the recombinant plasmid pNOV466 in which the nov-1 mutation was contained in a 3.1-kb insert. Experiments with pure plasmid DNA (Table 2), showed that pNOV466 was able to transform S. pneumoniae to novobiocin resistance (Nov R) at a high efficiency. Even higher levels of chromoso ...
... to the isolation of the recombinant plasmid pNOV466 in which the nov-1 mutation was contained in a 3.1-kb insert. Experiments with pure plasmid DNA (Table 2), showed that pNOV466 was able to transform S. pneumoniae to novobiocin resistance (Nov R) at a high efficiency. Even higher levels of chromoso ...
"Evolutionary History and Impact of Human DNA Transposons". In
... MITE consists of two 37-bp TIRs separated by 6 unique base pairs. Palindromes and inverted-repeat motifs are a known source of instability and DSBs in both prokaryotic and eukaryotic chromosomes, including those of mammals. It is important to note that even repeats relatively divergent in sequence o ...
... MITE consists of two 37-bp TIRs separated by 6 unique base pairs. Palindromes and inverted-repeat motifs are a known source of instability and DSBs in both prokaryotic and eukaryotic chromosomes, including those of mammals. It is important to note that even repeats relatively divergent in sequence o ...
Homework 7
... For this assignment you are required to have the following four class constants: • one for the minimum number of codons a valid protein must have, as an integer (default of 5) • a second for the percentage of mass from C and G in order for a protein to be valid, as an integer (default of 30) • a thi ...
... For this assignment you are required to have the following four class constants: • one for the minimum number of codons a valid protein must have, as an integer (default of 5) • a second for the percentage of mass from C and G in order for a protein to be valid, as an integer (default of 30) • a thi ...
spec
... For this assignment you are required to have the following four class constants: one for the minimum number of codons a valid protein must have, as an integer (default of 5) a second for the percentage of mass from C and G in order for a protein to be valid, as an integer (default of 30) a thi ...
... For this assignment you are required to have the following four class constants: one for the minimum number of codons a valid protein must have, as an integer (default of 5) a second for the percentage of mass from C and G in order for a protein to be valid, as an integer (default of 30) a thi ...
Slide 1
... (except for MonoZygotic twins) Therefore, individuals have small differences in nucleotide sequences of their DNA ...
... (except for MonoZygotic twins) Therefore, individuals have small differences in nucleotide sequences of their DNA ...
DNA and RNA Exam Questions (due: ) - A
... The diagram shows the bases in a rRNA molecule. In addition to A, U, C and G there are other bases present in tRNA. The other bases are shown by the letter D. ...
... The diagram shows the bases in a rRNA molecule. In addition to A, U, C and G there are other bases present in tRNA. The other bases are shown by the letter D. ...
Lab exam 1 V DONE
... same location, but four of them map to chromosome 4 and the remaining six map to chromosome 6. Other scientists get the same data as well (for the same species) and even confirm these loci encode for mRNA. Select the answer that explains these results. Select either D or E if you think there are two ...
... same location, but four of them map to chromosome 4 and the remaining six map to chromosome 6. Other scientists get the same data as well (for the same species) and even confirm these loci encode for mRNA. Select the answer that explains these results. Select either D or E if you think there are two ...
clicking here - PhysarumPlus
... the Dictyostelium database (http://dictybase.org), were extremely helpful and led to this suggested course of events: 1. Partner with the Dicty database, using their servers/hardware and "database schema". Rex suggests making cosmetic changes to the "look" of our site to clearly delineate the Physar ...
... the Dictyostelium database (http://dictybase.org), were extremely helpful and led to this suggested course of events: 1. Partner with the Dicty database, using their servers/hardware and "database schema". Rex suggests making cosmetic changes to the "look" of our site to clearly delineate the Physar ...
The chromosomal location of genes for elongation
... higher (74 YO amino acid identity) than the similarity between the Spirulina protein and that of other bacteria. A much lower degree of similarity (about 30% amino acid identity) exists between the cyanobacterial and the archaeal S 10 protein. The mutual relationships between the SlO sequences are c ...
... higher (74 YO amino acid identity) than the similarity between the Spirulina protein and that of other bacteria. A much lower degree of similarity (about 30% amino acid identity) exists between the cyanobacterial and the archaeal S 10 protein. The mutual relationships between the SlO sequences are c ...
Bio 6B Lecture Slides - D
... Primers & DNA Synthesis • DNA polymerases cannot initiate the synthesis of a polynucleotide. They can only add more nucleotides to the 3ʹ′ end of a present oligo- or poly-nucleotide. • The initial nucleotide strand to start is called a primer. – In cells, the primer is a 5–10-nucleotide RNAoligom ...
... Primers & DNA Synthesis • DNA polymerases cannot initiate the synthesis of a polynucleotide. They can only add more nucleotides to the 3ʹ′ end of a present oligo- or poly-nucleotide. • The initial nucleotide strand to start is called a primer. – In cells, the primer is a 5–10-nucleotide RNAoligom ...
AQ Molecular Genetics Unit Plan from V.Lee C.Mokono and J.Paas
... perform a diagnostic test for the ELL. It may be a simple oral check such as asking for a “thumbs up” from the students if they understand the concept (see example on day 3 of the unit overview) or the “1-2-3 strategy where the students can identify their level of understanding to the teacher. They ...
... perform a diagnostic test for the ELL. It may be a simple oral check such as asking for a “thumbs up” from the students if they understand the concept (see example on day 3 of the unit overview) or the “1-2-3 strategy where the students can identify their level of understanding to the teacher. They ...
Microsatellite

A microsatellite is a tract of repetitive DNA in which certain DNA motifs (ranging in length from 2–5 base pairs) are repeated, typically 5-50 times. Microsatellites occur at thousands of locations in the human genome and they are notable for their high mutation rate and high diversity in the population. Microsatellites and their longer cousins, the minisatellites, together are classified as VNTR (variable number of tandem repeats) DNA. The name ""satellite"" refers to the early observation that centrifugation of genomic DNA in a test tube separates a prominent layer of bulk DNA from accompanying ""satellite"" layers of repetitive DNA. Microsatellites are often referred to as short tandem repeats (STRs) by forensic geneticists, or as simple sequence repeats (SSRs) by plant geneticists.They are widely used for DNA profiling in kinship analysis and in forensic identification. They are also used in genetic linkage analysis/marker assisted selection to locate a gene or a mutation responsible for a given trait or disease.