Transfer of genetic material between the
... generally ,1 kb in length. This small amount of chloroplast DNA is in contrast to that found in the rice genome where the DNA from the rice chloroplast aligned with a total of between 780 000 and 933 600 bp in the DNA of the nuclear genome (International Rice Genome Sequencing Project, 2005). In our ...
... generally ,1 kb in length. This small amount of chloroplast DNA is in contrast to that found in the rice genome where the DNA from the rice chloroplast aligned with a total of between 780 000 and 933 600 bp in the DNA of the nuclear genome (International Rice Genome Sequencing Project, 2005). In our ...
Gel Electrophoresis
... Pulsed field gel electrophoresis DNA fragments longer than about 20 kb cannot be resolved in conventional agarose gel electrophoresis because long DNA molecules align themselves as rods and migrate with a mobility that is independent of their length. In pulsed field gel electrophoresis (PFGE), the mo ...
... Pulsed field gel electrophoresis DNA fragments longer than about 20 kb cannot be resolved in conventional agarose gel electrophoresis because long DNA molecules align themselves as rods and migrate with a mobility that is independent of their length. In pulsed field gel electrophoresis (PFGE), the mo ...
Novel Research Starts with GAPDH - Bio-Rad
... To identify differences in GAPDH code we must isolate plant DNA and amplify the gene of interest using PCR first with degenerate primers (primers that account for variation in the DNA code) ...
... To identify differences in GAPDH code we must isolate plant DNA and amplify the gene of interest using PCR first with degenerate primers (primers that account for variation in the DNA code) ...
Illustrating Python via Bioinformatics Examples
... Department of Informatics, University of Oslo ...
... Department of Informatics, University of Oslo ...
EPICENTRE Enzyme Catalog
... manufactures, and sells high-quality enzyme systems for life science research. Located in Madison, Wisconsin, EPICENTRE was founded in 1987, and now occupies a state-of-the art 72,000-s.f. building. EPICENTRE is well-known for its unique expertise in making a broad range of enzymes for molecular bio ...
... manufactures, and sells high-quality enzyme systems for life science research. Located in Madison, Wisconsin, EPICENTRE was founded in 1987, and now occupies a state-of-the art 72,000-s.f. building. EPICENTRE is well-known for its unique expertise in making a broad range of enzymes for molecular bio ...
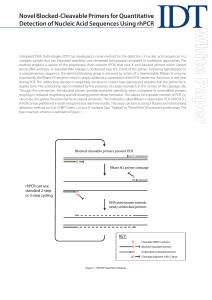
Novel Blocked-Cleavable Primers for Quantitative Detection of
... Improved Specificity: Discriminating Closely-Related Genes Human tumors are often grown in immune-deficit rodent systems as a routine part of oncology research. The human tumor mass becomes infiltrated with rodent fibroblasts and endothelial cells. Even though sequences differ between these xenogene ...
... Improved Specificity: Discriminating Closely-Related Genes Human tumors are often grown in immune-deficit rodent systems as a routine part of oncology research. The human tumor mass becomes infiltrated with rodent fibroblasts and endothelial cells. Even though sequences differ between these xenogene ...
Test Info Sheet
... Pontocerebellar hypoplasia (PCH) is a rare disorder affecting the ventral pons and cerebellum, two structures that share the same neuronal lineage during brain development. PCH has a fetal onset in most cases and appears to result from a combination of a developmental defect and progressive atrophy ...
... Pontocerebellar hypoplasia (PCH) is a rare disorder affecting the ventral pons and cerebellum, two structures that share the same neuronal lineage during brain development. PCH has a fetal onset in most cases and appears to result from a combination of a developmental defect and progressive atrophy ...
ANALYSIS OF MULTIPLE RESTRICTION FRAGMENT LENGTH
... duplicate nitrocellulose filters with 12p-labeled CRI cDNA probes . Probes. The cDNA probes, CRI-1 and CRI-2, are Eco RI fragments of CRI cDNA clone XT8 .3 that had been subdoned in pBR327 (17) . CRI-4 is the 381-bp Eco RI fragment between CRI-I and CR1-2 that was obtained from an overlapping phage ...
... duplicate nitrocellulose filters with 12p-labeled CRI cDNA probes . Probes. The cDNA probes, CRI-1 and CRI-2, are Eco RI fragments of CRI cDNA clone XT8 .3 that had been subdoned in pBR327 (17) . CRI-4 is the 381-bp Eco RI fragment between CRI-I and CR1-2 that was obtained from an overlapping phage ...
Rapid screening of PCR products using a novel agarose gel
... Fig 2. Comparison of separations on a large-format agarose gel vs. a Ready-To-Run gel. (A) 1 µl of a 1:10 dilution of PCR product separated in a 2% agarose gel containing SYBR Green I at 83 V for approximately 2 h. (B) 1 µl of a 1:5 dilution of PCR product separated in a 1.5% agarose Ready-To-Run ge ...
... Fig 2. Comparison of separations on a large-format agarose gel vs. a Ready-To-Run gel. (A) 1 µl of a 1:10 dilution of PCR product separated in a 2% agarose gel containing SYBR Green I at 83 V for approximately 2 h. (B) 1 µl of a 1:5 dilution of PCR product separated in a 1.5% agarose Ready-To-Run ge ...
Detection of genetically modified plants
... It must be sensitive and reliable enough to obtain exact results in all control laboratories. ...
... It must be sensitive and reliable enough to obtain exact results in all control laboratories. ...
A Retrospective Study of Balanced Chromosomal Translocations in
... The chromosomal disorders make a significant contribution to human mortality and morbidity. There are two kinds of chromosomal rearrangements: structural and numerical. Balanced translocations accepted as structural chromosomal abnormalities in humans commonly seen with a frequency of 1/600 (Van Dyk ...
... The chromosomal disorders make a significant contribution to human mortality and morbidity. There are two kinds of chromosomal rearrangements: structural and numerical. Balanced translocations accepted as structural chromosomal abnormalities in humans commonly seen with a frequency of 1/600 (Van Dyk ...
Review A model for chromosome structure during the mitotic
... matrix strands. This orientation is maintained into prophase when the nuclear matrix strand is converted into two closely associated sister chromatid cores with sister DNA loops extending in opposite directions. We propose that chromatid cores are contractile and show, using a physical model, that c ...
... matrix strands. This orientation is maintained into prophase when the nuclear matrix strand is converted into two closely associated sister chromatid cores with sister DNA loops extending in opposite directions. We propose that chromatid cores are contractile and show, using a physical model, that c ...
Alus
... and Alus • An estimated 500-2,000 Alu elements are restricted to the human genome. The vast majority of Alu insertions occur in non-coding regions and are thought to be evolutionarily neutral. However, an Alu insertion in the NF-1 gene is responsible for neurofibromatosis I, Alu insertions in intron ...
... and Alus • An estimated 500-2,000 Alu elements are restricted to the human genome. The vast majority of Alu insertions occur in non-coding regions and are thought to be evolutionarily neutral. However, an Alu insertion in the NF-1 gene is responsible for neurofibromatosis I, Alu insertions in intron ...
Fulltext PDF - Indian Academy of Sciences
... Analysis of root tip cells proved that both 31505 and 315051 had the chromosome number with 2n=42. Investigation of the meiosis showed that most observed cells of these two lines had 21 bivalents at meiotic MI, which indicated that these two lines were cytologically stable. GISH analysis was conduct ...
... Analysis of root tip cells proved that both 31505 and 315051 had the chromosome number with 2n=42. Investigation of the meiosis showed that most observed cells of these two lines had 21 bivalents at meiotic MI, which indicated that these two lines were cytologically stable. GISH analysis was conduct ...
View/Open
... fragment, without a centromere, and a centric fragment, with a centromere. The centric fragment migrates normally during the division process because it has a centromere.The acentric fragment, however, is soon lost. It is subsequently excluded from the nuclei formed and eventually degrades. In other ...
... fragment, without a centromere, and a centric fragment, with a centromere. The centric fragment migrates normally during the division process because it has a centromere.The acentric fragment, however, is soon lost. It is subsequently excluded from the nuclei formed and eventually degrades. In other ...
How imprinting is relevant to human disease - Development
... syndrome, Williams syndrome, Russell-Silver syndrome, etc. the possibility that they represent uniparental disomy for other chromosomes must be explored, since they are syndromes in which the major abnormalities consist of disharmonic growth and abnormal behaviour rather than major structural congen ...
... syndrome, Williams syndrome, Russell-Silver syndrome, etc. the possibility that they represent uniparental disomy for other chromosomes must be explored, since they are syndromes in which the major abnormalities consist of disharmonic growth and abnormal behaviour rather than major structural congen ...
Part III: Laboratory – Electrophoresis
... AGO1 and thus to locate the gene. The F2 plants that are homozygous for the mutation of interest (+/+), and thus showing the mutant phenotype, will be used for mapping. Since both of their chromosomes contain the mutation and the mutation was from a Ler background, the number of crossing-over events ...
... AGO1 and thus to locate the gene. The F2 plants that are homozygous for the mutation of interest (+/+), and thus showing the mutant phenotype, will be used for mapping. Since both of their chromosomes contain the mutation and the mutation was from a Ler background, the number of crossing-over events ...
Distinguishing Different DNA Heterozygotes by
... SNP of interest may or may not interfere with genotyping, depending on the analysis method (10 ). All PCR-based methods can be compromised if polymorphisms occur under the primers and lead to undesired allele-specific PCR. The same concern applies to internal primers used for sequencing or extension ...
... SNP of interest may or may not interfere with genotyping, depending on the analysis method (10 ). All PCR-based methods can be compromised if polymorphisms occur under the primers and lead to undesired allele-specific PCR. The same concern applies to internal primers used for sequencing or extension ...
Vitis 37 (3), 119
... The electrophoretically detected degree of genetic similarity between each pair of cultivars studied (Tab. 2) was calculated using the NTSYS-pc package 1.8 developed by ROHFL (Exeter Software, New York, USA). ...
... The electrophoretically detected degree of genetic similarity between each pair of cultivars studied (Tab. 2) was calculated using the NTSYS-pc package 1.8 developed by ROHFL (Exeter Software, New York, USA). ...
2- pcr primer design and reaction optimisation
... partly why one should not do more than about 30 amplification cycles: however, it is possible to reduce the denaturation temperature after about 10 rounds of amplification, as the mean length of target DNA is decreased: for templates of 300bp or less, denaturation temperature may be reduced to as lo ...
... partly why one should not do more than about 30 amplification cycles: however, it is possible to reduce the denaturation temperature after about 10 rounds of amplification, as the mean length of target DNA is decreased: for templates of 300bp or less, denaturation temperature may be reduced to as lo ...
Week 2. DNA isolation and PCR
... of PCR and DNA replication with a student from another group. Then, I lead a class discussion that begins with having the students volunteer similarities and differences between PCR and DNA replication. I like to show the PCR video (https://www.youtube.com/watch?v=2KoLnIwoZKU) in the laboratory and ...
... of PCR and DNA replication with a student from another group. Then, I lead a class discussion that begins with having the students volunteer similarities and differences between PCR and DNA replication. I like to show the PCR video (https://www.youtube.com/watch?v=2KoLnIwoZKU) in the laboratory and ...
Comparative genomic hybridization

Comparative genomic hybridization is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions (a portion of a whole chromosome). This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resoIution of 5-10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.This is achieved through the use of competitive fluorescence in situ hybridization. In short, this involves the isolation of DNA from the two sources to be compared, most commonly a test and reference source, independent labelling of each DNA sample with a different fluorophores (fluorescent molecules) of different colours (usually red and green), denaturation of the DNA so that it is single stranded, and the hybridization of the two resultant samples in a 1:1 ratio to a normal metaphase spread of chromosomes, to which the labelled DNA samples will bind at their locus of origin. Using a fluorescence microscope and computer software, the differentially coloured fluorescent signals are then compared along the length of each chromosome for identification of chromosomal differences between the two sources. A higher intensity of the test sample colour in a specific region of a chromosome indicates the gain of material of that region in the corresponding source sample, while a higher intensity of the reference sample colour indicates the loss of material in the test sample in that specific region. A neutral colour (yellow when the fluorophore labels are red and green) indicates no difference between the two samples in that location.CGH is only able to detect unbalanced chromosomal abnormalities. This is because balanced chromosomal abnormalities such as reciprocal translocations, inversions or ring chromosomes do not affect copy number, which is what is detected by CGH technologies. CGH does, however, allow for the exploration of all 46 human chromosomes in single test and the discovery of deletions and duplications, even on the microscopic scale which may lead to the identification of candidate genes to be further explored by other cytological techniques.Through the use of DNA microarrays in conjunction with CGH techniques, the more specific form of array CGH (aCGH) has been developed, allowing for a locus-by-locus measure of CNV with increased resolution as low as 100 kilobases. This improved technique allows for the aetiology of known and unknown conditions to be discovered.