Replication Animation Lab
... 9. Base pairing means that one strand is ___________ to the other strand. 10. What type of bond connects the two strands of DNA? ...
... 9. Base pairing means that one strand is ___________ to the other strand. 10. What type of bond connects the two strands of DNA? ...
Chapter 12-1 Skeleton Notes
... – Was it the DNA inside the bacteriophage or the protein coat surrounding the bacteriophage • Wanted to confirm Avery’s experiment Labeled the protein coat with a radioactive sulfur isotope and the DNA with a radioactive phosphorous isotope so that they may follow where each part goes after the inf ...
... – Was it the DNA inside the bacteriophage or the protein coat surrounding the bacteriophage • Wanted to confirm Avery’s experiment Labeled the protein coat with a radioactive sulfur isotope and the DNA with a radioactive phosphorous isotope so that they may follow where each part goes after the inf ...
DNA Notes How was the DNA Model Formed? 1) In the 1950`s a
... 2) Soon after a young chemist named Rosalind Franklin created images of DNA using a technique known as X-ray diffraction. Franklin used X rays to create images on film and found that the general shape of DNA is a spiral shape 3) In 1953 ___James_ ___Watson__, an American biochemist, and ___Francis__ ...
... 2) Soon after a young chemist named Rosalind Franklin created images of DNA using a technique known as X-ray diffraction. Franklin used X rays to create images on film and found that the general shape of DNA is a spiral shape 3) In 1953 ___James_ ___Watson__, an American biochemist, and ___Francis__ ...
Genetic Tools
... • Mr. and Mrs. Raider are deeply worried about their child who seems to be developing at a slower rate. They are concerned for the child’s health just like any other parent and have come to you for help. ...
... • Mr. and Mrs. Raider are deeply worried about their child who seems to be developing at a slower rate. They are concerned for the child’s health just like any other parent and have come to you for help. ...
Ch 13 Genetic Engineering
... • Scientists can synthesize a DNA strand and connect it to a circular DNA molecule known as a plasmid… which can be found naturally in bacteria. This bacteria can then be injected into a plant, and will insert its DNA into the plant. • If transformation is successful, the recombinant DNA is integra ...
... • Scientists can synthesize a DNA strand and connect it to a circular DNA molecule known as a plasmid… which can be found naturally in bacteria. This bacteria can then be injected into a plant, and will insert its DNA into the plant. • If transformation is successful, the recombinant DNA is integra ...
Document
... i. What would my sex chromosomes be if I were female? ______ ii. If I were male? ________ iii. Has anyone ever been born without an X chromosome? ______ 11. What is the purpose of a karyotype? a. List 3 things a karyotype shows? b. What does homologous chromosomes mean? 12. Human gametes contain ___ ...
... i. What would my sex chromosomes be if I were female? ______ ii. If I were male? ________ iii. Has anyone ever been born without an X chromosome? ______ 11. What is the purpose of a karyotype? a. List 3 things a karyotype shows? b. What does homologous chromosomes mean? 12. Human gametes contain ___ ...
PRE-AP Stage 3 – Learning Plan
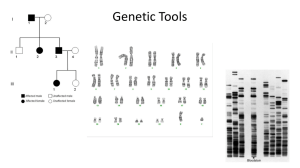
... Assessment and Closing: Exit ticket will be the final product of the pedigree chart that was created. Opening: Warm-up to review Pedigrees and Karyotypes Guided Practice: Karyotype Lab-Which disorder do you have based on the karyotype. New Material: DNA fingerprinting Assessment and Closing: Complet ...
... Assessment and Closing: Exit ticket will be the final product of the pedigree chart that was created. Opening: Warm-up to review Pedigrees and Karyotypes Guided Practice: Karyotype Lab-Which disorder do you have based on the karyotype. New Material: DNA fingerprinting Assessment and Closing: Complet ...
1 - web.biosci.utexas.edu
... 4. Which of the following contributes significantly to variation in nuclear genome size among plants. a. amounts of highly repetitive DNA b. amount of selfish DNA (e.g., such as transposons) c. frequency of introns d. a and b e. all of the above 5. Which of the following is incorrect concerning the ...
... 4. Which of the following contributes significantly to variation in nuclear genome size among plants. a. amounts of highly repetitive DNA b. amount of selfish DNA (e.g., such as transposons) c. frequency of introns d. a and b e. all of the above 5. Which of the following is incorrect concerning the ...
DNA Replication - The Biology Corner
... 5. The other side is the lagging strand - its moving away from the helicase (in the 5' to 3' direction). Problem: it reaches the replication fork, but the helicase is moving in the opposite direction. It stops, and another polymerase binds farther down the chain. This process creates several fragmen ...
... 5. The other side is the lagging strand - its moving away from the helicase (in the 5' to 3' direction). Problem: it reaches the replication fork, but the helicase is moving in the opposite direction. It stops, and another polymerase binds farther down the chain. This process creates several fragmen ...
C. Nucleic acid hybridization assays using cloned target DNA, and
... membrane. The labeled probe (whether by isotope or nonisotope labeling methods) is first denatured (made single strand) and then hybridized in solution to the nylon membrane containing the template DNA. After hybridization, the location of the fragments in the blotted template DNA that hybridized to ...
... membrane. The labeled probe (whether by isotope or nonisotope labeling methods) is first denatured (made single strand) and then hybridized in solution to the nylon membrane containing the template DNA. After hybridization, the location of the fragments in the blotted template DNA that hybridized to ...
DNA, Genes, and Chromosome Quiz
... 24.) DNA is converted into RNA during the process of DNA _____________________________________. This process occurs in the __________________________________. ****Bonus***** 1.) What is the 3 letter sequence that codes for an amino acid called? ...
... 24.) DNA is converted into RNA during the process of DNA _____________________________________. This process occurs in the __________________________________. ****Bonus***** 1.) What is the 3 letter sequence that codes for an amino acid called? ...
Genes - ASW Moodle
... A. When cells divide, they must make copies of the (wound up DNA) so that each cell receives a complete set of chromosomes *If the cell does not receive a complete set the cell is mutated and may not function ...
... A. When cells divide, they must make copies of the (wound up DNA) so that each cell receives a complete set of chromosomes *If the cell does not receive a complete set the cell is mutated and may not function ...
Gene Therapy
... How are the fragments separated? (by what trait) What is the end result? What are some uses of electrophoresis? ...
... How are the fragments separated? (by what trait) What is the end result? What are some uses of electrophoresis? ...
Genetics Online Scavenger Hunt
... What is DNA? 1. The instructions that provide all the information necessary for living organisms to grow and live are located in the ____________________. 2. The instructions come in the form of a molecule called ___________. 3. What do the letters in DNA stand for? _______________________________ 4 ...
... What is DNA? 1. The instructions that provide all the information necessary for living organisms to grow and live are located in the ____________________. 2. The instructions come in the form of a molecule called ___________. 3. What do the letters in DNA stand for? _______________________________ 4 ...
Within minutes, 2nd Generation ATP® tests answer the question
... microorganisms – can be difficult to manage thus making these processes difficult to troubleshoot. Through Microbe Detectives’ advanced metagenomics platform, nearly all microorganisms are identified, providing ground-breaking insight on optimization opportunities. ...
... microorganisms – can be difficult to manage thus making these processes difficult to troubleshoot. Through Microbe Detectives’ advanced metagenomics platform, nearly all microorganisms are identified, providing ground-breaking insight on optimization opportunities. ...
DNA Webquest L3
... The human chromosome is made of _______ total chromosomes ( two sets of ________). Where does each set come from? ___________________________________________________________________________ About ______% of the human genome has no known purpose. The remaining 3% that does have a known purpose is ...
... The human chromosome is made of _______ total chromosomes ( two sets of ________). Where does each set come from? ___________________________________________________________________________ About ______% of the human genome has no known purpose. The remaining 3% that does have a known purpose is ...
In meiosis, what is the difference between metaphase 1 and
... Try to read over the review and the end of each chapter. Again- important highlights. Also, on Yves web page is the test 2 “outline” which mentions some end of chapter questions that are relevant to the test/lectures. Give ‘em a try. The answers for these are in the back of the book. Sleep well, eat ...
... Try to read over the review and the end of each chapter. Again- important highlights. Also, on Yves web page is the test 2 “outline” which mentions some end of chapter questions that are relevant to the test/lectures. Give ‘em a try. The answers for these are in the back of the book. Sleep well, eat ...
DNA fingerprinting
... 6. The binding of the probe is visualised using radioactivity, fluorescence, conjugated enzyme. 7. The resulting band patterns are a fingerprint. 8. The final DNA fingerprint is built by using several probes (5-10 or more) simultaneously. ...
... 6. The binding of the probe is visualised using radioactivity, fluorescence, conjugated enzyme. 7. The resulting band patterns are a fingerprint. 8. The final DNA fingerprint is built by using several probes (5-10 or more) simultaneously. ...
Supplementary Methods
... visualisation and score were carried out as described5. AFLP analysis: AFLP analysis was performed as described6 with slight modifications7. Preamplification was carried out using EcoRI + A / MseI + C primers. Two primer combinations were used for selective amplification: EcoRI + ACT / MseI + CAG an ...
... visualisation and score were carried out as described5. AFLP analysis: AFLP analysis was performed as described6 with slight modifications7. Preamplification was carried out using EcoRI + A / MseI + C primers. Two primer combinations were used for selective amplification: EcoRI + ACT / MseI + CAG an ...
Reg Bio DNA tech 2013 ppt
... Useful for: person’s paternity, identifying human remains, tracing human origins, and providing evidence in a criminal case. 98% of genetic makeup doesn’t code for proteins Compare segments that vary the most from person to person (noncoding segments that repeat over and over) ...
... Useful for: person’s paternity, identifying human remains, tracing human origins, and providing evidence in a criminal case. 98% of genetic makeup doesn’t code for proteins Compare segments that vary the most from person to person (noncoding segments that repeat over and over) ...
AP Biology
... 9. Why do scientists use a radioactive isotope tag for the probes? 10. How is DNA denaturation different than protein denaturation? ...
... 9. Why do scientists use a radioactive isotope tag for the probes? 10. How is DNA denaturation different than protein denaturation? ...
Comparative genomic hybridization

Comparative genomic hybridization is a molecular cytogenetic method for analysing copy number variations (CNVs) relative to ploidy level in the DNA of a test sample compared to a reference sample, without the need for culturing cells. The aim of this technique is to quickly and efficiently compare two genomic DNA samples arising from two sources, which are most often closely related, because it is suspected that they contain differences in terms of either gains or losses of either whole chromosomes or subchromosomal regions (a portion of a whole chromosome). This technique was originally developed for the evaluation of the differences between the chromosomal complements of solid tumor and normal tissue, and has an improved resoIution of 5-10 megabases compared to the more traditional cytogenetic analysis techniques of giemsa banding and fluorescence in situ hybridization (FISH) which are limited by the resolution of the microscope utilized.This is achieved through the use of competitive fluorescence in situ hybridization. In short, this involves the isolation of DNA from the two sources to be compared, most commonly a test and reference source, independent labelling of each DNA sample with a different fluorophores (fluorescent molecules) of different colours (usually red and green), denaturation of the DNA so that it is single stranded, and the hybridization of the two resultant samples in a 1:1 ratio to a normal metaphase spread of chromosomes, to which the labelled DNA samples will bind at their locus of origin. Using a fluorescence microscope and computer software, the differentially coloured fluorescent signals are then compared along the length of each chromosome for identification of chromosomal differences between the two sources. A higher intensity of the test sample colour in a specific region of a chromosome indicates the gain of material of that region in the corresponding source sample, while a higher intensity of the reference sample colour indicates the loss of material in the test sample in that specific region. A neutral colour (yellow when the fluorophore labels are red and green) indicates no difference between the two samples in that location.CGH is only able to detect unbalanced chromosomal abnormalities. This is because balanced chromosomal abnormalities such as reciprocal translocations, inversions or ring chromosomes do not affect copy number, which is what is detected by CGH technologies. CGH does, however, allow for the exploration of all 46 human chromosomes in single test and the discovery of deletions and duplications, even on the microscopic scale which may lead to the identification of candidate genes to be further explored by other cytological techniques.Through the use of DNA microarrays in conjunction with CGH techniques, the more specific form of array CGH (aCGH) has been developed, allowing for a locus-by-locus measure of CNV with increased resolution as low as 100 kilobases. This improved technique allows for the aetiology of known and unknown conditions to be discovered.