Chapter 15 – Recombinant DNA and Genetic Engineering
... – In bacterial cells – Insert foreign DNA (gene) into and put back in bacteria – reproduces naturally making a DNA clone – Cloning vector: plasmid used to accept foreign DNA and replicate it • Reverse transcriptase: enzyme from RNA viruses that perform transcription in reverse (RNA to DNA) – cDNA: ( ...
... – In bacterial cells – Insert foreign DNA (gene) into and put back in bacteria – reproduces naturally making a DNA clone – Cloning vector: plasmid used to accept foreign DNA and replicate it • Reverse transcriptase: enzyme from RNA viruses that perform transcription in reverse (RNA to DNA) – cDNA: ( ...
BSC 219
... Due 10/18/12 1) ( 3 points) Describe the main ways that eukaryotic transcription initiation is different from prokaryotic transcription initiation. Eukaryotic initiation involves a large number of proteins to form an initiation complex that recruits RNA Polymerase to the promoter region. The DNA seq ...
... Due 10/18/12 1) ( 3 points) Describe the main ways that eukaryotic transcription initiation is different from prokaryotic transcription initiation. Eukaryotic initiation involves a large number of proteins to form an initiation complex that recruits RNA Polymerase to the promoter region. The DNA seq ...
D: Glossary of Acronyms and Terms
... Natural selection: The process of differential reproductive success by which genes in a population increase or decrease in frequency with the passage of generations, depending on their contribution to the survival of offspring in which they are carried; arguably the most important of the several mec ...
... Natural selection: The process of differential reproductive success by which genes in a population increase or decrease in frequency with the passage of generations, depending on their contribution to the survival of offspring in which they are carried; arguably the most important of the several mec ...
The Dinosaur Heresies
... source of evolutionary novelty, especially in bacteria Modularity may not be the primary mode of innovation in protein sequences ...
... source of evolutionary novelty, especially in bacteria Modularity may not be the primary mode of innovation in protein sequences ...
Digitally Programmed Cells
... • Collaboration with Ron Breaker / Adam Roth • Discovery of unique riboswitches specific for GTP rather than dGTP • Found in no other sequenced genomes ...
... • Collaboration with Ron Breaker / Adam Roth • Discovery of unique riboswitches specific for GTP rather than dGTP • Found in no other sequenced genomes ...
Restriction-Modification Systems as Minimal Forms of Life
... A restriction (R) endonuclease recognizes a specific DNA sequence and introduces a double-strand break (Fig. 1A). A cognate modification (M) enzyme methylates the same sequence and thereby protects it from cleavage. Together, these two enzymes form a restriction-modification system. The genes encodi ...
... A restriction (R) endonuclease recognizes a specific DNA sequence and introduces a double-strand break (Fig. 1A). A cognate modification (M) enzyme methylates the same sequence and thereby protects it from cleavage. Together, these two enzymes form a restriction-modification system. The genes encodi ...
Do plants have more genes than humans?
... wheat or rice and corn, it appears that intergenic regions have expanded by insertions of retrotransposons, and that larger genomes contain more junk DNA between genes7. Interestingly, repetitive DNA between genomes of sorghum and maize do not cross-hybridize, although they have some common retrotra ...
... wheat or rice and corn, it appears that intergenic regions have expanded by insertions of retrotransposons, and that larger genomes contain more junk DNA between genes7. Interestingly, repetitive DNA between genomes of sorghum and maize do not cross-hybridize, although they have some common retrotra ...
Human Genome
... Have students conduct this experiment to see, feel, and smell DNA. 1. Measure 2/3 cup of dried split peas. Add about 1/8 teaspoon of salt and 1-1/3 cups of cold water. Mix the ingredients in a blender on high speed for 15 seconds. 2. Pour the pea mixture through a strainer into another container. Ad ...
... Have students conduct this experiment to see, feel, and smell DNA. 1. Measure 2/3 cup of dried split peas. Add about 1/8 teaspoon of salt and 1-1/3 cups of cold water. Mix the ingredients in a blender on high speed for 15 seconds. 2. Pour the pea mixture through a strainer into another container. Ad ...
Document
... complex as ssRNAs and initiate destruction of all cellular RNAs that share homology to the dsRNA. RNAi has been incredibly useful to researchers because it can be used to reduce the expression of genes that are tough to mutate. TFIID is a complex of proteins within the basal/general transcriptional ...
... complex as ssRNAs and initiate destruction of all cellular RNAs that share homology to the dsRNA. RNAi has been incredibly useful to researchers because it can be used to reduce the expression of genes that are tough to mutate. TFIID is a complex of proteins within the basal/general transcriptional ...
Ross - Tree Improvement Program
... • Two copies are identical = “homozygous” • Two copies are different = “heterozygous” Homozygous parent ...
... • Two copies are identical = “homozygous” • Two copies are different = “heterozygous” Homozygous parent ...
Lecture slides
... – knowledge-based • Chou-Fasman and GOR methods for SSE prediction • Comparative modeling and protein threading for tertiary structure prediction ...
... – knowledge-based • Chou-Fasman and GOR methods for SSE prediction • Comparative modeling and protein threading for tertiary structure prediction ...
No Slide Title
... Guo and Kemphues, Cell 81, 611 (1995) observed that sense and antisense strands worked equally at reducing transcript, – in an anti-sense experiment, a gene is constructed so that it produces a complementary strand to an expressed transcript, • the goal is to complement, thus inactivate the mRNA. ...
... Guo and Kemphues, Cell 81, 611 (1995) observed that sense and antisense strands worked equally at reducing transcript, – in an anti-sense experiment, a gene is constructed so that it produces a complementary strand to an expressed transcript, • the goal is to complement, thus inactivate the mRNA. ...
Fact sheet (PDF, 58.54 KB) (opens in a new window)
... Little is known regarding the impact of transcriptional interference on gene expression. Researchers at the University of Western Sydney in collaboration with the University of New South Wales have developed a novel method to both detect and regulate transcriptional interference between genes of int ...
... Little is known regarding the impact of transcriptional interference on gene expression. Researchers at the University of Western Sydney in collaboration with the University of New South Wales have developed a novel method to both detect and regulate transcriptional interference between genes of int ...
gene families
... have been innumerable paracentric inversions within the arms, but very few pericentric inversions that would mix the arms, and relatively few translocations or transpositions between different chromosomes. The autosomal arms themselves have not even been reassociated with each other, e.g. 2L and 2R ...
... have been innumerable paracentric inversions within the arms, but very few pericentric inversions that would mix the arms, and relatively few translocations or transpositions between different chromosomes. The autosomal arms themselves have not even been reassociated with each other, e.g. 2L and 2R ...
poster in ppt
... It has been engineered in order to be used into Swarm or others agent based simulation's models, to easy obtain "minded" agents who are fully autonomous, able to decide their own behaviors and able to change it to fit in different environmental conditions. Another main usage of the algorithm is to s ...
... It has been engineered in order to be used into Swarm or others agent based simulation's models, to easy obtain "minded" agents who are fully autonomous, able to decide their own behaviors and able to change it to fit in different environmental conditions. Another main usage of the algorithm is to s ...
Biological Agents Special Edition of eBulletin
... be considered where the potential for super-Mendelian inheritance is lost in subsequent generations (eg due to segregation of system components). Finally, if alternatives cannot be used, and autonomous systems are planned, additional containment and control measures may be required. These will vary ...
... be considered where the potential for super-Mendelian inheritance is lost in subsequent generations (eg due to segregation of system components). Finally, if alternatives cannot be used, and autonomous systems are planned, additional containment and control measures may be required. These will vary ...
GenomicVariation_11-22
... Rather than look at multiple, different regulatory regions from one species, look at one region but across multiple, orthologous regions from many species. Hypothesis: functional regions of the genome will be conserved more than ‘nonfunctional’ regions, due to selection. Therefore, simply look for r ...
... Rather than look at multiple, different regulatory regions from one species, look at one region but across multiple, orthologous regions from many species. Hypothesis: functional regions of the genome will be conserved more than ‘nonfunctional’ regions, due to selection. Therefore, simply look for r ...
The divergence of duplicate genes in Arabidopsis
... outgroup • >30 showed significantly unequal rates of evolution • no evident chromosomal or regional biases Distance measure ...
... outgroup • >30 showed significantly unequal rates of evolution • no evident chromosomal or regional biases Distance measure ...
REGULATION OF GENE EXPRESSION IN EUKARYOTES
... further required to regulate the activity of gene expression ...
... further required to regulate the activity of gene expression ...
Genetics and genomics in wildlife studies: Implications for
... of extant biodiversity. There is a continuing need for theoretical, statistical, and bioinformatic developments because of the huge amount of NGS information becoming available. For example, genome-wide association studies (GWAS) have had mixed success in accounting for the total heritability of tra ...
... of extant biodiversity. There is a continuing need for theoretical, statistical, and bioinformatic developments because of the huge amount of NGS information becoming available. For example, genome-wide association studies (GWAS) have had mixed success in accounting for the total heritability of tra ...
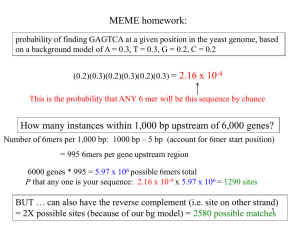
GENETICS - St. Bonaventure University
... and use a figure of 3,000 bp per gene then we need only 75 million bp for all our genes. We have about 3.2 billion base pairs in our DNA!! ...
... and use a figure of 3,000 bp per gene then we need only 75 million bp for all our genes. We have about 3.2 billion base pairs in our DNA!! ...
here - CMBI
... • First group in-paralogs in every species • Find bi-directional best hits between inparalogous groups • Join in-paralogs to orthologous groups – Link all pairs of in-paralogous groups – Only if link is confirmed by third species (triangle) ...
... • First group in-paralogs in every species • Find bi-directional best hits between inparalogous groups • Join in-paralogs to orthologous groups – Link all pairs of in-paralogous groups – Only if link is confirmed by third species (triangle) ...
Transposable element
A transposable element (TE or transposon) is a DNA sequence that can change its position within the genome, sometimes creating or reversing mutations and altering the cell's genome size. Transposition often results in duplication of the TE. Barbara McClintock's discovery of these jumping genes earned her a Nobel prize in 1983.TEs make up a large fraction of the C-value of eukaryotic cells. There are at least two classes of TEs: class I TEs generally function via reverse transcription, while class II TEs encode the protein transposase, which they require for insertion and excision, and some of these TEs also encode other proteins. It has been shown that TEs are important in genome function and evolution. In Oxytricha, which has a unique genetic system, they play a critical role in development. They are also very useful to researchers as a means to alter DNA inside a living organism.