Reconciling the many faces of lateral gene transfer
... within bacterial genomes impart distinctive biases to the composition of long-term residents of the genome [10], such that recently acquired genes will appear aberrant by comparison if they have evolved in a genome with different mutational biases. All such methods rely on a robust description of wh ...
... within bacterial genomes impart distinctive biases to the composition of long-term residents of the genome [10], such that recently acquired genes will appear aberrant by comparison if they have evolved in a genome with different mutational biases. All such methods rely on a robust description of wh ...
Print as PDF
... gene sets of interest. They can be used to visualize bipartite clusters (Hierarchical Similarity [HiSim] Graph), or visualize genes with the more common intersections, GeneSet Graph. Generation and visualization of a maximal triclique using the intersection of gene sets with the Triclique Viewer Too ...
... gene sets of interest. They can be used to visualize bipartite clusters (Hierarchical Similarity [HiSim] Graph), or visualize genes with the more common intersections, GeneSet Graph. Generation and visualization of a maximal triclique using the intersection of gene sets with the Triclique Viewer Too ...
escienceweekkaty
... – Bergen Center for Computational Science – Gene Prediction in Algal Viruses, a case study. • Workflow assembles evidence for predicted genes / potential functions • Human expert can ‘review’ this evidence before submission to the genome database ...
... – Bergen Center for Computational Science – Gene Prediction in Algal Viruses, a case study. • Workflow assembles evidence for predicted genes / potential functions • Human expert can ‘review’ this evidence before submission to the genome database ...
Genome browsers for power users
... already have) just as before (NOT via genome graph) • Then, import this set into the genome graph tool (under custom tracks) • What is your impressions? Where are they? ...
... already have) just as before (NOT via genome graph) • Then, import this set into the genome graph tool (under custom tracks) • What is your impressions? Where are they? ...
Bioinformatics - Sequences and Computers
... Language and DNA use sequences to communicate information. The sequence elements in language are letters and punctuation, in DNA they are the nucleotides. As the letters in books contain information that is realized by readers, the sequence of nucleotides in DNA contains information that is realized ...
... Language and DNA use sequences to communicate information. The sequence elements in language are letters and punctuation, in DNA they are the nucleotides. As the letters in books contain information that is realized by readers, the sequence of nucleotides in DNA contains information that is realized ...
Guide to using the PCR lab File
... certain regions of the genome. When assembling the genome sequence, long segments on one chromosome appeared to be identical to regions elsewhere in the genome. Such regions are called segmental duplications. These duplicated segments are complete copies of entire segments of DNA sequence with all t ...
... certain regions of the genome. When assembling the genome sequence, long segments on one chromosome appeared to be identical to regions elsewhere in the genome. Such regions are called segmental duplications. These duplicated segments are complete copies of entire segments of DNA sequence with all t ...
011 Chapter 11 Microbial Genetics: Gene Structure Replication amp
... 54. Cellular RNA molecules are always initially single stranded, but they often have double stranded regions because of internal base pairing. True False 55. Split genes have been found in both procaryotes and eucaryotes but are much more common in eucaryotes. True False 56. The basic differences be ...
... 54. Cellular RNA molecules are always initially single stranded, but they often have double stranded regions because of internal base pairing. True False 55. Split genes have been found in both procaryotes and eucaryotes but are much more common in eucaryotes. True False 56. The basic differences be ...
genotyping single nucleotide polymorphisms located on
... time of flight mass spectrometry (MALDI-TOF MS) due to its inherent speed and accuracy for typing SNPs. The speed and accuracy of MALDI-TOF MS also allows rapid development of large DNA typing databases and population studies. Because SNPs are typically bi-alleic, a greater number of these markers a ...
... time of flight mass spectrometry (MALDI-TOF MS) due to its inherent speed and accuracy for typing SNPs. The speed and accuracy of MALDI-TOF MS also allows rapid development of large DNA typing databases and population studies. Because SNPs are typically bi-alleic, a greater number of these markers a ...
Bioinformatics and drug target selection for malaria control
... open the ability to test the efficacy of drugs in robust model systems prior to clinical trials. The challenge of identifying good drug targets will rely on integration of disparate data from high-throughput technologies such as genome and cDNA sequencing, microarrays, proteomics, structural genomic ...
... open the ability to test the efficacy of drugs in robust model systems prior to clinical trials. The challenge of identifying good drug targets will rely on integration of disparate data from high-throughput technologies such as genome and cDNA sequencing, microarrays, proteomics, structural genomic ...
Gene Expression Specific Target Amplification
... The BioMark™ System uses a sample loading volume of 5 µL, and distributes this sample mixture across 48 or 96 reaction chambers in 9 or 6 nL aliquots, respectively. With these micro-volumes, detecting the specific targets requires a minimum of 500-1,000 copies in the original 5 µL loading volume. Be ...
... The BioMark™ System uses a sample loading volume of 5 µL, and distributes this sample mixture across 48 or 96 reaction chambers in 9 or 6 nL aliquots, respectively. With these micro-volumes, detecting the specific targets requires a minimum of 500-1,000 copies in the original 5 µL loading volume. Be ...
Annotation
... the appropriate place. Always change the output format to coordinates. Use L5 (or closest choice) as the coding potential model. The RBS(ribosomal binding site) model is M. tuberculosis (is annotating a Mycobacteriophage). All of these can be deselcted (and should be) and run. Print the output. This ...
... the appropriate place. Always change the output format to coordinates. Use L5 (or closest choice) as the coding potential model. The RBS(ribosomal binding site) model is M. tuberculosis (is annotating a Mycobacteriophage). All of these can be deselcted (and should be) and run. Print the output. This ...
An Introduction to MaizeGDB
... Integrated help throughout the site, including tutorial BLAST tool which provides map locations of hits Easy-to-use advanced searches Tab-delimited search results which you can download to your own analysis programs Tools integrated into retrieved records. Extensive linking to other resources. ...
... Integrated help throughout the site, including tutorial BLAST tool which provides map locations of hits Easy-to-use advanced searches Tab-delimited search results which you can download to your own analysis programs Tools integrated into retrieved records. Extensive linking to other resources. ...
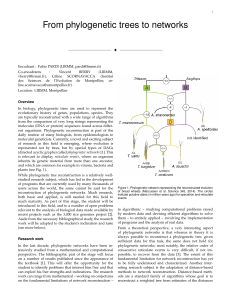
From phylogenetic trees to networks
... represented not by trees, but by special types of DAGs (directed acyclic graphs) called phylogenetic networks [1]. This is relevant to display reticulate events, where an organism inherits its genetic material from more than one ancestor, and which are common for example in viruses, bacteria and pla ...
... represented not by trees, but by special types of DAGs (directed acyclic graphs) called phylogenetic networks [1]. This is relevant to display reticulate events, where an organism inherits its genetic material from more than one ancestor, and which are common for example in viruses, bacteria and pla ...
1 Genome Project-write: A Grand Challenge Using Synthesis, Gene
... Because of the special challenges surrounding human genomes, this activity will include an expanded ELSI component. It will also be explicitly limited to work in cells, and organoids derived from them only. ...
... Because of the special challenges surrounding human genomes, this activity will include an expanded ELSI component. It will also be explicitly limited to work in cells, and organoids derived from them only. ...
Lecture 4 Genome_Organization
... • Some genes don’t have any introns. Most common example is the histone genes. Histones are the proteins DNA gets wrapped around in the lowest unit of chromosomal organization, the nucleosome. • Some genes are quite huge: dystrophin (associated with Duchenne muscular dystrophy) is 2.4 Mbp and takes ...
... • Some genes don’t have any introns. Most common example is the histone genes. Histones are the proteins DNA gets wrapped around in the lowest unit of chromosomal organization, the nucleosome. • Some genes are quite huge: dystrophin (associated with Duchenne muscular dystrophy) is 2.4 Mbp and takes ...
Long Noncoding RNAs May Alter Chromosome`s 3D
... from West Africa, signaling a secThe result is a mosaic of DNA ond wave of slave trade from what segments with different histories. is now Cameroon and Congo, Such admixtures complicate parsaccording to Moreno-Estrada. Genetic heritage. The mixed ancestry of Caribbean people is ing out the genetic g ...
... from West Africa, signaling a secThe result is a mosaic of DNA ond wave of slave trade from what segments with different histories. is now Cameroon and Congo, Such admixtures complicate parsaccording to Moreno-Estrada. Genetic heritage. The mixed ancestry of Caribbean people is ing out the genetic g ...
Syllabus Science Microbiology
... c. Work of Winogradsky and Beijerinck. Discovery of microorganisms as plant pathogens. 2. The Microbial World (10 Hours) a. Distribution of microorganisms in nature. b. Diversity in microbial habitat. Types of microorganisms. c. Introduction to prokaryotic world, eukaryotic microorganisms, viruses a ...
... c. Work of Winogradsky and Beijerinck. Discovery of microorganisms as plant pathogens. 2. The Microbial World (10 Hours) a. Distribution of microorganisms in nature. b. Diversity in microbial habitat. Types of microorganisms. c. Introduction to prokaryotic world, eukaryotic microorganisms, viruses a ...
DNA Chip Analysis and Bioinformatics
... Paste the probe DNA sequence into the query box, scroll down and select “show results in a new window” and click “ BLAST”. Leave all other parameters as they are. 6. Wait until the page loads (this could take a minute or so - be patient). 7. Scroll down to “Sequences producing significant alignments ...
... Paste the probe DNA sequence into the query box, scroll down and select “show results in a new window” and click “ BLAST”. Leave all other parameters as they are. 6. Wait until the page loads (this could take a minute or so - be patient). 7. Scroll down to “Sequences producing significant alignments ...
Homework - The Fenyo Lab
... D) Load the BED file and the BAM file into IGV, zoom in and take a screenshot of the reads around MACS_peak_10, 11, and 12. And then look at MACS_peak_13. Scroll left and right a few kb at a time. Why are there aligned reads all over the genome? Why do you think there is no peak called near chr1:1, ...
... D) Load the BED file and the BAM file into IGV, zoom in and take a screenshot of the reads around MACS_peak_10, 11, and 12. And then look at MACS_peak_13. Scroll left and right a few kb at a time. Why are there aligned reads all over the genome? Why do you think there is no peak called near chr1:1, ...
Full text for subscribers
... research endeavours by the animal biotechnologists striving to analyse single-nucleotide polymorphisms “SNPs” among genes and DNA markers are also helping to improve breeding strategies. Recently, the advent of next generation sequencing (NGS) technology allowed de novo sequencing of the goat genome ...
... research endeavours by the animal biotechnologists striving to analyse single-nucleotide polymorphisms “SNPs” among genes and DNA markers are also helping to improve breeding strategies. Recently, the advent of next generation sequencing (NGS) technology allowed de novo sequencing of the goat genome ...
Teacher notes and student sheets
... or carefully matched on all the other factors that might have an effect. To assess the outcome of a cohort study, scientists compare the number of cases in the two groups after a period of exposure. To judge that a factor does affect the outcome, the difference must be big enough not to be attributa ...
... or carefully matched on all the other factors that might have an effect. To assess the outcome of a cohort study, scientists compare the number of cases in the two groups after a period of exposure. To judge that a factor does affect the outcome, the difference must be big enough not to be attributa ...
... actual open reading frame responsible remains unknown. Among these are several temperature-sensitive lethal mutations known as unknown (Inoue and Ishikawa, 1970; Ishikawa and Perkins, 1983). As part of our continuing effort to define the gene defect associated with these otherwise anonymous temperat ...
Sieracki_lecture1_july6 - C-MORE
... Two Flavobacterium with proteorhodopsin genes. These are being whole genome sequenced by JGI. There may be a PCR bias against Flavobacteria (Kirchman, et al. 2000) ...
... Two Flavobacterium with proteorhodopsin genes. These are being whole genome sequenced by JGI. There may be a PCR bias against Flavobacteria (Kirchman, et al. 2000) ...
Metagenomics

Metagenomics is the study of genetic material recovered directly from environmental samples. The broad field may also be referred to as environmental genomics, ecogenomics or community genomics. While traditional microbiology and microbial genome sequencing and genomics rely upon cultivated clonal cultures, early environmental gene sequencing cloned specific genes (often the 16S rRNA gene) to produce a profile of diversity in a natural sample. Such work revealed that the vast majority of microbial biodiversity had been missed by cultivation-based methods. Recent studies use either ""shotgun"" or PCR directed sequencing to get largely unbiased samples of all genes from all the members of the sampled communities. Because of its ability to reveal the previously hidden diversity of microscopic life, metagenomics offers a powerful lens for viewing the microbial world that has the potential to revolutionize understanding of the entire living world. As the price of DNA sequencing continues to fall, metagenomics now allows microbial ecology to be investigated at a much greater scale and detail than before.