* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Recombinant protein expression in E.coli
Epigenetics in learning and memory wikipedia , lookup
Genome evolution wikipedia , lookup
Designer baby wikipedia , lookup
Gene nomenclature wikipedia , lookup
Gene expression programming wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Non-coding RNA wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Non-coding DNA wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genomic library wikipedia , lookup
History of genetic engineering wikipedia , lookup
Gene expression profiling wikipedia , lookup
Protein moonlighting wikipedia , lookup
Primary transcript wikipedia , lookup
DNA vaccination wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Helitron (biology) wikipedia , lookup
Point mutation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
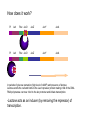
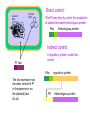
Recombinant protein expression in E.coli Bio4600 2003 Vigdis Lauvrak Modern Biotechnology- enabling technologies Protein technologies Computational technologies Bioinformatics Folding Prediction Docking Homology Modelling Cloning Expression Purification Molecular Evolution Interaction Maps Structural Biology Crystallization Data Collection Structure Determination Host: E.coli Vector: Plasmid A tremendous number of modified strains A tremendous number of highly specialized constructs Growth medium Selectable marker gene E.coli replicon Promoter Plasmid DNA Cytoplasma Cloningsites Leader sequences Genomic DNA Tags Tags Gene to be expressed Periplasma Inner membrane Outer membrane Major options to be considered: •Gene dosage (copy number) •Level of expression •Which compartment to harvest from •Tags for purification, improvement of stability and solubility •Codon usage E.coli:recombinant protein •Purpose of expression: Large scale industrial/or analytical levels? A replicom is a genetic unit consisting of an origin of DNA replication and its associated elements. origin pBR322 colE1 pUC pMOB45 pACYC pSC101 Replicon pMB1 colE1 mod pMB1 pKN402 p15A pSC101 copy number 15-20 15-20 500-700 15-118 18-22 5 Gene dosage Medium to high copy number plasmids •Relaxed replication •Random distribution •Relatively low loss: Continously growth and toxic genes/gene products will lead to plasmid loss. Increased plasmid stability: Selectable markers •Genes for antibiotic resistance •Complementation: An essential chromosomal gene is deleted or mutated and an intact copy or a supressor is suplied in trans. •Genes or repressors that lead to cell death upon plasmid loss. Duplication of genomic inserts Increased gene dosage in E-coli genome: •RecA dupllication of insert (Olson et al. 1998) : 15--40 copies (may be unstable without a selectable marker). •Tn1545 site specific recombination (Peredelchuck and Bennett 1997) - time consuming Control of expression level Desired: High expression level (10-30% or more of produced protein) Observed: Many proteins may are toxic at high doses. Solution: Regulation of expression Basic elements of E.coli expression systems R P -35 SD coding sequence -10 TTGACA(N)17TATAAT TT STOP codon START codon mRNAUAAGGAGG(N)8AUG (91%) GUG (8%) UUG (1%) R: Reprossor P: Promoter SD: Shine Delgarno sequence (Ribosome binding site- start of mRNA) (TT: terminator (stabilizes mRNA)) UAU UGA UAG E.coli expression vectors: contain: • E.coli expression elements •Unique cloning sites •An origin of replication •A selectable marker Level of regulation depends on the promoter •The lac operon- the paradigm of protein regulation in E.coli: lactose/ IPTG-induction (derepression) •lacUV5 (leaky): IPTG •tac and trc synthetic versions of lac (tighter): IPTG •T7-late promoter : Depends on T7 polymerase •PL promoter- Lambda CI regulated, tight regulation •cspA: Cold chock induction •phoA, trp and araBAD (PBAD): Nutritional inducible •tet: Tetracycline inducible •Signal dependent promoters: pH, oxygen conc., osmolarity etc. (Inexpensive large scale production) The lac operon -the paradigm of expression regulation in E.coli lacI operon Pl lacI lac repressor Pl lacI lac Operon Plac lacO lacZ lacY Beta-galactosidase (cleavage of lactose) Plac lacO lacZ Beta galpermease (import of lactose) lacY lacA Beta galtransacetylase (function?) lacA In presence of glucose (no starvation/ low cAMP level) the lac repressor (lacI gene product) is bound to the lac operator and blocks RNA polymerase from binding DNA - Thus the lacI geneproduct acts as an repressor (inhibitor of transcription). In the absence How does it work? Pl lacI Plac lacO lacZ lacY lacA Pl lacI Plac lacO lacZ lacY lacA In periods of glucose starvation (high level of cAMP) and presence of lactose: Lactose enetrs the cell and binds to the LacI repressor protein making it fall of the DNA. RNA polymerase can now bind to the lac promoter and initiate transcription. -Lactose acts as an inducer (by removing the repressor) of transcription. The lac promoter of E.coli expression vectors: • Induction is performed with IPTG which acts as a synthetic lactose analogue that binds the lacI gene product. •Presence of glucose further prevents transcription from the lac promoter. IPTG Pl lacI PX lacO Pl lacI PX lacO The CI binding site (lac operator) can be combinde with various other promoter sequences to give improved regulation. Genomic DNA Plasmid DNA Direct control: Plac/PI may directly control the production of plasmid encoded heterologous protein: Plac Heterologous protein Indirect control: PI lacI A regualtory protein under lacI control Plac The lac repressor may be under control of PI in the genome or on the plasmid (lacIE.coli). PX regulatory protein Heterologous protein The pET 11 vectors (Novagen and Stratagene) with T7/lacO promoter : lacUV5 T7 polymerase •T7 RNA polymerase in the bacterial chromosome is controled by a lacUV5 promoter. • The heterologous protein is under control of the T7 promoter. •The T7 promoter is fused to the plac operator •The lac I repressor inhibits expression of T7 polymerase and the heterologous protein. •IPTG will induce is used for induction. T7 lacO Heterologous protein T7 terminator PI lacI A copy of the lacI gene (also found in the genome) is inserted on the plasmids to achieve sufficient repressor. Choice of E.coli compartment Growth medium Plasmid DNA Cytoplasma Genomic DNA Periplasma Inner membrane Outer membrane Cytoplasmic expression Advantages: •No need for signal sequences, •High concentration of expressed protein Disadvantages: •Formation of inclusion bodies •(No disulfide bond formation), •Protein instability, Periplasm Advantages •Improved folding (no inclusion body formation) •Disulfide bridge formation (may be enhanced by the presence of DsbA and DsbB proteins) •Fewer proteins and possible leakage to growth medium may facilitate purification. •Less protein degradation. Disadvantages. •Low protein concentration due to inefficient transport and small compartment Solution •Thight regulation of expression •Molecular chaperones (protein specific) •Temperature down shift after induction- less formation of inclusion bodies). Growth media No efficient system for direct transport to growth media. Leakage from periplasm is often used. Common problems encountered with E.coli expression system: The desired protein may be: Unstable, toxic, insoluble, form inclusion bodies, uncorect folded, depend on disulfide bridges, and active only with postranslational modifications : glycosylation, phosphorylation and amidation. Solutions: Choice of a suitable E.coli strain, tags, fusions and leader sequences can solve many problems including disulfide bridge formation, but proteins that need correct postranslational modifications as underlined above have to be produced in Eucaryotic systems. Solutions: •Thight regulation of expression •Coexpression of molecular chaperones (protein specific) •Reduction of rate of protein synthesis (lower growth rate by temperature down shift after induction) •Fusion moiteties may increase folding, solubility and resistance to proteolysis. •Use of protease deficient E.coli strains •Use of thioredoxin reductase (trxB) og glutatione reductase (gor) double mutants may give disulfide bridge formation in cytosol •Periplasmic expression Characteristics of suitable induction sensitive promotors •High strength •Tight regulation •Simple and cost effective induction: Basic research: IPTG (lactose analogue (toxic)) Tetracycline Thermal Industrial production of theraeutics: Thermal Chemical Nutrional