* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Tutorial_9_NEW
Hammerhead ribozyme wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Human genome wikipedia , lookup
Metagenomics wikipedia , lookup
Gene expression profiling wikipedia , lookup
Designer baby wikipedia , lookup
Long non-coding RNA wikipedia , lookup
X-inactivation wikipedia , lookup
Transposable element wikipedia , lookup
History of genetic engineering wikipedia , lookup
Non-coding DNA wikipedia , lookup
Microevolution wikipedia , lookup
Polyadenylation wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Helitron (biology) wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Epitranscriptome wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Primary transcript wikipedia , lookup
History of RNA biology wikipedia , lookup
RNA interference wikipedia , lookup
Nucleic acid tertiary structure wikipedia , lookup
Non-coding RNA wikipedia , lookup
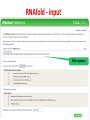
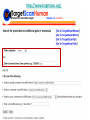
Tutorial 9 RNA Structure Prediction RNA Structure Prediction • RNA secondary structure prediction RNAfold, RNAalifold • microRNA prediction TargetScan – Cool story of the day: How viruses use miRNAs to attack humans RNA secondary structure prediction GGGCUAUUAGCUCAGUUGGUUA GAGCGCACCCCUGAUAAGGGUGA GGUCGCUGAUUCGAAUUCAGCAU AGCCCA Base pair probability RNA structure prediction by Vienna RNA package RNAfold server Minimum free energy structures and base pair probabilities from single RNA or DNA sequences. RNAalifold server Consensus secondary structures from an alignment of several related RNA or DNA sequences. You need to upload an alignment. http://rna.tbi.univie.ac.at/ RNAfold • Gives best stabilized structure (structure with minimal free energy (MFE)) • Uses a dynamic programming algorithm that exploits base pairing and thermodynamic probabilities in order to predict the most likely structures of an RNA molecule. RNAfold - input RNA sequence RNAfold - output Minimal free energy structure Structure prediction Free energy of the ensamble Best “average” structure Graphic representation MFE structure An average, may not exist in the ensemble RNAalifold Structure prediction based on alignments Alignment RNAalifold - output Understanding the color scheme C-G C-G U-A C-G C-G C-G G-C U-A A-U C-C http://www.almob.org/content/pdf/1748-7188-6-26.pdf MicroRNAs miRNA gene mature miRNA Target gene MicroRNA in Cancer Sun et al, 2012 The challenge for Bioinformatics: - Identifying new microRNA genes - Identifying the targets of specific microRNA How to find microRNA genes? Searching for sequences that fold to a hairpin ~70 nt -RNAfold -other efficient algorithms for identifying stem loops Concentrating on intragenic regions and introns - Filtering coding regions Filtering out non conserved candidates -Mature and pre-miRNA is usually evolutionary conserved How to find microRNA genes? A. Structure prediction B. Evolutionary Conservation Predicting microRNA targets MicroRNA targets are located in 3’ UTRs, and complementing mature microRNAs •Why is it hard to find them ?? – Base pairing is required only in the seed sequence (7-8 nt) – Lots of known miRNAs have similar seed sequences Very high probability to find by chance mature miRNA 3’ UTR of Target gene Predicting microRNA target genes • General methods - Find motifs which complements the seed sequence (allow mismatches) – Look for conserved target sites – Consider the MFE of the RNA-RNA pairing ∆G (miRNA+target) – Consider the delta MFE for RNA-RNA pairing versus the folding of the target ∆G (miRNA+target )- ∆G (target) http://www.targetscan.org/ Sum of phylogenetic branch lengths between species that contain a site More negative scores represent a more favorable site The stability of of a miRNAtarget duplex A score reflecting the probability that a site is conserved due to selective maintenance of miRNA targeting rather than by chance or any other reason. Mir 136 Mir 136 - conserved* microRNA * conserved across most mammals, but usually not beyond placental mammals How to evaluate our results True positive (TP) = correctly identified (miRNA targets correctly identified) False positive (FP)= incorrectly identified (non miRNA targets miss identified as targets) True negative (TN)= correctly rejected (non miRNA targets correctly not identified) False negative (FN) = incorrectly rejected (miRNA targets not identified) Sensitivity (recall)= True positive Rate = TP /(TP+FN) Specificity (precision)= True Negative Rate = TN /(TN+FP) Cool Story of the day How viruses use miRNAs to attack humans? The group developed an algorithm for predicting miRNA targets and applied it to human Cytomegalovirus (hcmv) miRNAs MICB, an immunorelated gene, was among the highest ranking predicted targets and the top prediction for hcmv-miR-UL112. They found that hcmv-miR-UL112 specifically down-regulates MICB expression during viral infection, leading to reduced killing by Natural Killer cells (A human virus-defense mechanism) Natural Killer (NK) cells • NK cells are cytotoxic lymphocyte that kill virus-infected cells and tumor cells. • In order to function they should be activated through receptors. One of these is NKG2D. • MICB is a stress-induced ligand of NK cells through the NKG2D receptor). Cerwenka et al. Nature Reviews Immunology 2001