* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Gene expression - Weizmann Institute of Science
Long non-coding RNA wikipedia , lookup
Transcription factor wikipedia , lookup
Minimal genome wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Point mutation wikipedia , lookup
Microevolution wikipedia , lookup
Gene expression profiling wikipedia , lookup
Genome (book) wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
History of genetic engineering wikipedia , lookup
Oncogenomics wikipedia , lookup
Designer baby wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Primary transcript wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
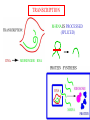
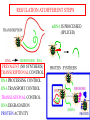
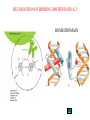
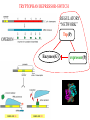
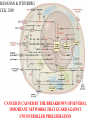
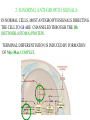
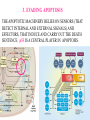
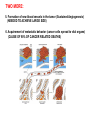
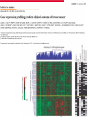
GENE EXPRESSION: (EUCARYOTIC) •GENE =SEGMENT OF DNA = BLUEPRINT FOR A SINGLE PROTEIN •EACH CELL CONTAINS ALL GENES !!! •WHEN A GENE IS EXPRESSED, THE PROTEIN IT CODES FOR IS SYNTHESIZED (AT RIBOSOMES) •NOT ALL GENES ARE EXPRESSED IN ALL CELLS (OF A MULTICELLULAR ORGANISM) OR AT ALL TIMES DIFFERENTIATION • DIFFERENT CELL TYPES SYNTHESIZE DIFFERENT PROTEINS • TYPICAL HUMAN CELL – ABOUT 10-20,000 EXPRESSED GENES • FOR THE 2000 MOST ABUNDANT (>50,000 COPIES/CELL) FEW DIFFERENCES ARE FOUND (5 FOLD OR LESS) • A FEW % ARE CELL TYPE SPECIFIC – BUT PROFILES DO DIFFER SIGNIFICANTLY A PARTICULAR CELL’S EXPRESSED GENES VARY •WITH TIME •IN RESPONSE TO EXTERNAL SIGNALS •IN RESPONSE TO INTERNAL “CLOCKS” •DEPENDING ON STATE – NORMAL, STRESSED, ABNORMAL HOW IS GENE EXPRESSION REGULATED?? TRANSCRIPTION M-RNA IS PROCESSED (SPLICED) REGULATION AT DIFFERENT STEPS mRNA IS PROCESSED (SPLICED) PREVALENT (NO SYNTHESIS) TRANSCRIPTIONAL CONTROL RNA PROCESSING CONTROL RNA TRANSPORT CONTROL TRANSLATIONAL CONTROL RNA DEGRADATION PROTEIN ACTIVITY TRANSCRIPTION – CLOSER LOOK RNA POLYMERASE ATTACHES TO DNA, MOVES ALONG IT, OPENS DOUBLE HELIX, SYNTHESIZES mRNA. CONTROL OF EXPRESSION BY ASSISTING OR BLOCKING ATTACHMENT OF RNA POLYMERASE TO DNA. RNA POLYMERASE BINDS AT A SPECIFIC REGION, THE PROMOTER, AT THE START OF THE GENE, IF (1) AN ACTIVATOR IS ATTACHED AT AN ADJACENT SPECIFIC REGULATORY BINDING SEQUENCE (OPERATOR) AND (2) A REPRESSOR IS NOT ATTACHED TO ITS OWN OPERATOR TRANSCRIPTION START TRANSCRIPTION – CLOSER LOOK RNA POLYMERASE DOES NOT BIND AT THE PROMOTER, AT THE START OF THE GENE, IF A REPRESSOR IS ATTACHED TO IT’S OWN OPERATOR TRANSCRIPTION FACTORS RECOGNITION OF BINDING MOTIFS IN DNA RECOGNITION OF BINDING MOTIFS IN DNA 2 HOMEODOMAIN TRYPTOPHAN REPRESSOR-SWITCH REGULATORY “NETWORK” Trp(P) OPERON Enzymes(G) repressor(P) lac OPERON – E-COLI (PROCARYOTIC) OPERON – SET OF GENES PLACED ONE AFTER THE OTHER ON DNA, TAKING PART IN ONE PROCESS (BREAKDOWN OF lactose). E-COLI PREFERS glucose – WILL PROCESS lactose ONLY UNDER ( –glucose/+ lactose ) CONDITIONS. 4-SWITCH! Cyclic AMP allolactose CONCENTRATION CONCENTRATION ACTIVATOR: REPRESSOR: --glucose/+lactose + + WHEN +glucose WHEN +lactose (ACTIVE IF COMPLEX) (ACTIVE IF FREE) Decision-making by bacteria: choosing between two sugars can be tough… The first detailed map of a gene’s decision-making computation (Setty, Alon, 2003) EUCARYOTIC – MUCH MORE COMPLEX • RNA POLYMERASE CANNOT INITIATE TRANSCRIPTION GENERAL TRANSCRIPTION FACTORS ASSEMBLE, FORM COMPLEX ON OPERATOR NEAR PROMOTER - ABUNDANT TBP – SUBUNIT OF TFIID • SPECIFIC REGULATORS OF TRANSCRIPTION (ENHANCERS) CAN ATTACH TO DNA MANY 1000 OF BP UPSTREAM, CAN EVEN BE PLACED DOWNSTREAM FROM START SITE VERY MINUTE AMOUNT PRESENT • MAY NEED MORE THAN ONE TRANSCRIPTION FACTOR TO ACTIVATE GENE proximity of GAL4 enhances 1000 fold the attachment of TFIIB to TFIID control of human beta-globin REGULATORY NETWORKS • TRANSCRIPTION FACTORS ARE PROTEINS THAT ACTIVATE OR REPRESS GENES’ TRANSCRIPTION INTO PROTEINS • PROTEINS FORM COMPLEXES THAT INDUCE /TURN OFF /REGULATE A GENE’S TRANSCRIPTIONAL CAPACITY COMPLEX NETWORKS OF REGULATION OF GENE EXPRESSION EMERGE HANAHAN & WEINBERG CELL 2000 CANCER IS CAUSED BY THE BREAKDOWN OF SEVERAL IMPORTANT NETWORKS,THAT GUARD AGAINST UNCONTROLLED PROLIFERATION NORMAL CELL STATES & CELL CYCLE G1 –gap, decide whether to proliferate, wait or cross to non-dividing stage G0 S -- DNA Synthesis G2– gap, allow DNA repair M – Mitosis, cell division Check Points (Internal and External signals) NORMAL ENTRY TO/EXIT FROM CELL CYCLE Programmed Cell Death (Apoptosis) Induced Apoptosis Growth Signals Cell Division Proliferation Limited replication, senescence, crisis too many divisions Cell cycle arrest (G0) or Terminal differentiation Anti Growth Signals Check Point (Internal and External signals) Cancer Cell-HALLMARKS series of random genetic accidents Programmed Cell Death (Apoptosis) 3 Cell Division Proliferation Limited replication, senescence, crisis 3 4 2 1 Cell cycle arrest or Terminal differentiation Defective computation at check points, or failure to interpret signals or execute instructions: 1 Proliferation becomes independent of growth factors. 2 Loosing responses to cell cycle inhibitory signals. 3 Failure to apoptose when necessary. 4 Immortalization. These are the main 4 HALLMARKS OF CANCER. 1. SELF SUFFICIENCY IN GROWTH SIGNALS IN NORMAL CELLS, GROWTH FACTORS ARE RELEASED BY NEIGHBOR CELLS, BOUND BY GF RECEPTORS (USUALLY IN CELL MEMBRANE), WHICH GET MODIFIED AND INITIATE A CASCADE OF SIGNALING EVENTS . What can go wrong? 1. Autonomous generation of growth factors 2. Receptor overexpression or alteration 3. Defective downstream processing MUTANT Ras SEND DOWNSTREAM GROWTH SIGNALS WITH NO STIMULUS FROM UPSTREAM 2. IGNORING ANTI-GROWTH SIGNALS IN NORMAL CELLS, MOST ANTIGROWTH SIGNALS DIRECTING THE CELL TO G0 ARE CHANNELED THROUGH THE Rb (RETINOBLASTOMA) PROTEIN. TERMINAL DIFFERENTIATION IS INDUCED BY FORMATION OF Myc-Max COMPLEX. 3. EVADING APOPTOSIS THE APOPTOTIC MACHINERY RELIES ON SENSORS (THAT DETECT INTERNAL AND EXTERNAL SIGNALS) AND EFFECTORS, THAT INDUCE AND CARRY OUT THE DEATH SENTENCE. p53 IS A CENTRAL PLAYER IN APOPTOSIS. 4. IMMORTALIZATION A CELL CAN UNDERGO A LIMITED NUMBER OF DIVISIONS. THE “COUNTING DEVICE” IS A STRING OF SEVERAL 1000 REPEATS OF A 6-BP SEQUENCE ELEMENT AT THE END OF THE CHROMOSOMES (TELOMERS). IN EACH DIVISION 50 – 100 TELOMERIC BP ARE LOST. WHEN THEY RUN OUT, THE CHROMOSOME ENDS ARE UNPROTECTED AND FUSE, LEADING TO CRISIS AND DEATH OF THE CELL. CANCER CELLS ACQUIRE TELOMERE MAINTENANCE Cancer Cell-HALLMARKS series of random genetic accidents Programmed Cell Death (Apoptosis) 3 Cell Division Proliferation Limited replication, senescence, crisis 3 4 2 1 Cell cycle arrest or Terminal differentiation Defective computation at check points, or failure to interpret signals or execute instructions: 1 Proliferation becomes independent of growth factors. 2 Loosing responses to cell cycle inhibitory signals. 3 Failure to apoptose when necessary. 4 Immortalization. These are the main 4 HALLMARKS OF CANCER. TWO MORE: 5. Formation of new blood vessels in the tumor (Sustained Angiogenesis) (NEEDED TO ACHIEVE LARGE SIZE) 6. Acquirement of metastatic behavior (cancer cells spread to vital organs) (CAUSE OF 90% OF CANCER RELATED DEATHS) Cancer results from anomalies in genes regulating cell growth: mutations, translocations • Two classes of genes are involved: • 1) Oncogenes - positive regulators promote cancer by hyperactivity (one allele is enough) • 2) Tumor Suppressor genes - negative regulators, promote cancer by loss of activity (both alleles must be mutated) AND/OR GENOMIC INSTABILITY: SCIENCE 2002 GENES ONCOGENES = GAS PEDAL TUMOR SUPPRESSORS = BRAKE PEDAL GENOME INTEGRITY GENES = MECHANIC AIMS: UNDERSTAND THE GENETIC/MOLECULAR MECHANISMS THAT CAUSE CANCER. WHICH PATHWAYS BREAK DOWN IN VARIOUS CANCERS, AND HOW? IDENTIFY SUB-CLASSES OF EACH DISEASE. CLINICAL RELEVANCE: PERSONALIZED PREDICTIVE PREVENTIVE MEDICINE: PREDICT OUTCOME, DESIGN SUITABLE THERAPY DRUG DISCOVERY MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia Armstrong et al, Nature Genetics 30, 41-47 (2002) PCA Leukemia 2 Korsmeyer ALL,AML,MLL 500 most separating genes GLEEVEC IN CML: