* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Introduction to Bioinformatics
Synthetic biology wikipedia , lookup
RNA silencing wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
Non-coding RNA wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Genome evolution wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Non-coding DNA wikipedia , lookup
Epitranscriptome wikipedia , lookup
Proteolysis wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Expanded genetic code wikipedia , lookup
Gene expression wikipedia , lookup
Protein structure prediction wikipedia , lookup
Deoxyribozyme wikipedia , lookup
List of types of proteins wikipedia , lookup
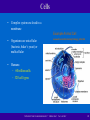
Nucleic acid analogue wikipedia , lookup
Biochemistry wikipedia , lookup
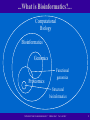
Introduction to Bioinformatics Doç. Dr. Nizamettin AYDIN [email protected] “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 1 Recommended Texts www.amazon.com “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 2 Recommended Texts - 2 www.amazon.com “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 3 Recommended Texts - 3 “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 4 Recommended Texts - 4 Bioinformatics for Dummies Jean Claverie, Cedric Notredame Bioinformatics: A Practical Guide to the Analysis of Genes and Proteins Andreas D. Baxevanis, B. F. Ouellette, Ouellette B. F. Francis. Instant Notes in Bioinformatics D. R. Westhead, Richard M. Twyman, J. H. Parish Bioinformatics: Sequence and Genome Analysis, Vol. 5 David W. Mount, David Mount Developing Bioinformatics Computer Skills Cynthia Gibas, Per Jambeck, Lorrie LeJeune (Editor) Discovering Genomics, Proteomics, and Bioinformatics A. Malcolm Campbell, Laurie J. Heyer “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 5 Recommended Texts - 5 Structural Bioinformatics Philip E. Bourne (Editor), Helge Weissig Beginning Perl for Bioinformatics James Tisdall Mastering Perl for Bioinformatics James D. Tisdall “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 6 What is Bioinformatics?... “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 7 ...What is Bioinformatics?... Computational Biology Bioinformatics Genomics Proteomics Functional genomics Structural bioinformatics “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 8 ...What is Bioinformatics? • Bioinformatics: collection and storage of biological information • Computational biology: development of algorithms and statistical models to analyze biological data • Bioinformatics/Computational Biology will be interchanged “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 9 Why is Bioinformatics Important? • Applications areas include – – – – – – – – Medicine Pharmaceutical drug design Toxicology Molecular evolution Biosensors Biomaterials Biological computing models DNA computing “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 10 Why should I care? • SmartMoney ranks Bioinformatics as #1 among next HotJobs • Business Week 50 Masters of Innovation • Jobs available, exciting research potential • Important information waiting to be decoded! “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 11 Why is bioinformatics hot? • Supply/demand: few people adequately trained in both biology and computer science • Genome sequencing, microarrays, etc lead to large amounts of data to be analyzed • Leads to important discoveries • Saves time and money “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 12 The Role of Computational Biology Source: GenBank GenBank BASEPAIR GROWTH 3.841 Millions 4.000 3.500 3.000 2.009 2.500 2.000 1.160 1.500 1.000 652 1 2 3 5 10 16 24 35 49 72 101 157 217 385 500 0 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 3D Structures Growth: Source: http://www.rcsb.org/pdb/ holdings.html “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 13 Fighting Human Disease • Genetic / Inherited – Diabetes • Viral – Flu, common cold • Bacterial – Meningitis, Strep throat “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 14 Drug Development Life Cycle Discovery (2 to 10 Years) Preclinical Testing (Lab and Animal Testing) Phase I (20-30 Healthy Volunteers used to check for safety and dosage) Phase II (100-300 Patient Volunteers used to check for efficacy and side effects) Phase III (1000-5000 Patient Volunteers used to monitor reactions to long-term drug use) $600-700 Million! FDA Review & Approval Post-Marketing Testing Years 0 2 4 6 8 10 12 14 16 7 – 15 Years! “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 15 Drug lead screening 5,000 to 10,000 compounds screened 5 Drug Candidates enter Clinical Testing; 250 Lead Candidates in Preclinical Testing 80% Pass Phase I 30%Pass Phase II 80% Pass Phase III One drug approved by the FDA “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 16 What skills are needed? • Well-grounded in one of the following areas: – Computer science – Molecular biology – Statistics • Working knowledge and appreciation in the others! “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 17 Where Can I Learn More? • • • • • ISCB: http://www.iscb.org/ NBCI: http://ncbi.nlm.nih.gov/ http://www.bioinformatics.org/ Journals Conferences “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 18 Overview of Molecular Biology • • • • • • • Cells Chromosomes DNA RNA Amino Acids Proteins Genome/Transcriptome/Proteome “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 19 Cells • Complex system enclosed in a membrane Example Animal Cell • Organisms are unicellular (bacteria, baker’s yeast) or multicellular www.ebi.ac.uk/microarray/ biology_intro.htm • Humans: – 60 trillion cells – 320 cell types “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 20 Organisms • Classified into two types: • Eukaryotes: contain a membrane-bound nucleus and organelles (plants, animals, fungi,…) • Prokaryotes: lack a true membrane-bound nucleus and organelles (single-celled, includes bacteria) • Not all single celled organisms are prokaryotes! “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 21 Chromosomes • In eukaryotes, nucleus contains one or several double stranded DNA molecules organized as chromosomes Human Karyotype http://avery.rutgers.edu/WSSP/StudentScholars/ Session8/Session8.html • Humans: – 22 Pairs of autosomes – 1 pair sex chromosomes “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 22 Chromosomes Image source: www.biotec.or.th/Genome/whatGenome.html “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 23 DNA is the blueprint for life • DNA: Deoxyribonucleic Acid • Every cell in your body has 23 chromosomes in the nucleus • The genes in these chromosomes determine all of your physical attributes. • Single stranded molecule (oligomer, polynucleotide) chain of nucleotides • 4 different nucleotides: – – – – Adenosine (A) Cytosine (C) Guanine (G) Thymine (T) “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 24 Mapping the Genome • The human genome project has provided us with a draft of the entire human genome. • Four bases: A, T, C, G • 3.12 billion basepairs • 99% of these are the same • Polymorphisms = where they differ “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 25 Nucleotide Bases • Purines (A and G) • Pyrimidines (C and T) • Difference is in base structure Image Source: www.ebi.ac.uk/microarray/ biology_intro.htm “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 26 DNA • Can be thought of as an alphabet with 4 characters • 4 letter alphabet with sufficiently long words contains information to create complex organisms • Not unlike a computer with a small alphabet “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 27 DNA polynucleotides(oligomers) • Different nucleotides are strung together to form polynucleotides • Ends of the polynucleotide are different • A directionality is present • Convention is to label the coding strand from 5’ to 3’ http://www.emc.maricopa.edu/faculty/farabee/BIOBK/BioBookDNAMOLGEN.html “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 28 Single Strand Polynucleotide Example polynucleotide: 5’ GTAAAGTCCCGTTAGC 3’ “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 29 Double Stranded DNA • DNA can be single-stranded or double-stranded • Double stranded DNA: second strand is the “reverse complement” strand • Reverse complement runs in opposite direction and bases are complementary • Complementary bases: – A, T – C, G “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 30 Double Stranded Sequence Example double stranded polynucleotide: 5’ GTAAAGTCCCGTTAGC 3’ | | | | | | | | | | | | | | | | 3’ CATTTCAGGGCAATCG 5’ http://www.emc.maricopa.edu/faculty/farabee/BIOBK/BioBookDNAMOLGEN.html “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 31 Double Stranded DNA “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 32 Double Helix • Two complementary DNA strands form a stable DNA double helix • This spring marks the 50th anniversary of its discovery Image source; www.ebi.ac.uk/microarray/ biology_intro.htm “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 33 “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 34 How does the code work? • Template for construction of proteins “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 35 Proteins: Molecular machinery • Proteins in your muscles allows you to move: myosin and actin “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 36 Proteins: Molecular machinery • Enzymes (digestion, catalysis) • Structure (collagen) “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 37 Proteins: Molecular machinery • Signaling (hormones, kinases) • Transport (energy, oxygen) Image source: Crane digital, http://www.cranedigital.com/ “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 38 Example Case: HIV Protease 1. Exposure & infection 2. HIV enters your cell 3. Your own cell reads the HIV “code” and creates the HIV proteins. 4. New viral proteins prepare HIV for infection of other cells. © George Eade, Eade Creative Services, Inc. http://whyfiles.org/035aids/index.html “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 39 HIV Protease & Inhibition “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 40 HIV Protease as a drug target • Many drugs bind to protein active sites. • This HIV protease can no longer prepare HIV proteins for infection, because an inhibitor is already bound in its active site. HIV Protease + Peptidyl inhibitor (1A8G.PDB) “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 41 Drug Discovery • Target Identification – What protein can we attack to stop the disease from progressing? • Lead discovery & optimization – What sort of molecule will bind to this protein? • Toxicology – Does it kill the patient? – Does it have side effects? – Does it get to the problem spots? “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 42 Drug discovery: past & present • Put some of the infectious agent into thousands of tiny wells • Add a known drug lead compound into each well. – Try nearly every drug lead known. • See which ones kill the agent… – Too small to see, so we have to use chemical tests called assays “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 43 Finding drug leads • Once we have a target, how do we find some compounds that might bind to it? • The old way: exhaustive screening • The new way: computational screening! “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 44 Drug Lead Screening & Docking ? • Complementarity – Shape – Chemical – Electrostatic “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 45 Problems in Bioinformatcs • Genomics – Gene finding – Annotation • Sequence alignment and database search – Functional genomics • Microarray expression, “gene chips” • Proteomics – Structure prediction • Comparative modeling – Function prediction • Structural bioinformatics – Molecular docking, screening, etc. “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 46 RNA • Ribonucleic Acid • Similar to DNA • Thymine (T) is replaced by uracil (U) • RNA can be: – Single stranded – Double stranded – Hybridized with DNA “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 47 RNA • RNA is generally single stranded • Forms secondary or tertiary structures • RNA folding will be discussed later • Important in a variety of ways, including protein synthesis “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 48 RNA secondary structure • E. coli Rnase P RNA secondary structure “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 49 mRNA • Messenger RNA • Linear molecule encoding genetic information copied from DNA molecules • Transcription: process in which DNA is copied into an RNA molecule “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 50 mRNA processing • Eukaryotic genes can be pieced together – Exons: coding regions – Introns: non-coding regions • mRNA processing removes introns, splices exons together • Processed mRNA can be translated into a protein sequence “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 51 mRNA Processing Image source: http://departments.oxy.edu/biology/Stillman/bi221/111300/processing_of_hnrnas.htm “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 52 tRNA • Transfer RNA • Well-defined three-dimensional structure • Critical for creation of proteins “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 53 tRNA structure “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 54 tRNA • Amino acid attached to each tRNA • Determined by 3 base anticodon sequence (complementary to mRNA) • Translation: process in which the nucleotide sequence of the processed mRNA is used in order to join amino acids together into a protein with the help of ribosomes and tRNA “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 55 Genetic Code • • • • • • • 4 possible bases (A, C, G, U) 3 bases in the codon 4 * 4 * 4 = 64 possible codon sequences Start codon: AUG Stop codons: UAA, UAG, UGA 61 codons to code for amino acids (AUG as well) 20 amino acids – redundancy in genetic code “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 56 20 Amino Acids • • • • • • • • • • • • • • • • • • • • • • Glycine (G, GLY) Alanine (A, ALA) Valine (V, VAL) Leucine (L, LEU) Isoleucine (I, ILE) Phenylalanine (F, PHE) Proline (P, PRO) Serine (S, SER) Threonine (T, THR) Cysteine (C, CYS) Methionine (M, MET) Tryptophan (W, TRP) Tyrosine (T, TYR) Asparagine (N, ASN) Glutamine (Q, GLN) Aspartic acid (D, ASP) Glutamic Acid (E, GLU) Lysine (K, LYS) Arginine (R, ARG) Histidine (H, HIS) START: AUG STOP: UAA, UAG, UGA “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 57 Amino Acids • building blocks for proteins (20 different) • vary by side chain groups • Hydrophilic amino acids are water soluable • Hydrophobic are not • Linked via a single chemical bond (peptide bond) • Peptide: Short linear chain of amino acids (< 30) polypeptide: long chain of amino acids (which can be upwards of 4000 residues long). “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 58 Proteins • Polypeptides having a three dimensional structure. • Primary–sequence of amino acids constituting the polypeptide chain • Secondary–local organization into secondary structures such as helices and sheets • Tertiary –three dimensional arrangements of the amino acids as they react to one another due to the polarity and resulting interactions between their side chains • Quaternary–number and relative positions of the protein subunits “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 59 Protein Structure “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 60 Central Dogma DNA RNA PROTEIN “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 61 Central Dogma “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 62 What is a Gene? • the physical and functional unit of heredity that carries information from one generation to the next • DNA sequence necessary for the synthesis of a functional protein or RNA molecule “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 63 Genome • chromosomal DNA of an organism • number of chromosomes and genome size varies quite significantly from one organism to another • Genome size and number of genes does not necessarily determine organism complexity “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 64 Genome Comparison ORGANISM CHROMOSOMES GENOME SIZE GENES Homo sapiens (Humans) 23 3,200,000,000 ~ 30,000 Mus musculus (Mouse) 20 2,600,000,000 ~30,000 Drosophila melanogaster (Fruit Fly) 4 180,000,000 ~18,000 Saccharomyces cerevisiae (Yeast) 16 14,000,000 ~6,000 Zea mays (Corn) 10 2,400,000,000 ??? “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 65 Transcriptome • complete collection of all possible mRNAs (including splice variants) of an organism. • regions of an organism’s genome that get transcribed into messenger RNA. • transcriptome can be extended to include all transcribed elements, including non-coding RNAs used for structural and regulatory purposes. “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 66 Proteome • the complete collection of proteins that can be produced by an organism. • can be studied either as static (sum of all proteins possible) or dynamic (all proteins found at a specific time point) entity “INTRODUCTION TO BIOINFORMATICS” “SPRING 2005” “Dr. N AYDIN” 67