* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Lecture slides
Gene therapy wikipedia , lookup
Gene desert wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Ridge (biology) wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Gene nomenclature wikipedia , lookup
Molecular ecology wikipedia , lookup
Genetic engineering wikipedia , lookup
Metabolomics wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Expression vector wikipedia , lookup
Biosynthesis wikipedia , lookup
Real-time polymerase chain reaction wikipedia , lookup
Point mutation wikipedia , lookup
Gene expression wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Metabolic network modelling wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Pharmacometabolomics wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Biochemical cascade wikipedia , lookup
Gene regulatory network wikipedia , lookup
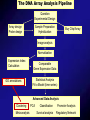
Analysis of GO annotation at cluster level by Agnieszka S. Juncker The DNA Array Analysis Pipeline Question Experimental Design Array design Probe design Sample Preparation Hybridization Buy Chip/Array Image analysis Normalization Expression Index Calculation GO annotations Comparable Gene Expression Data Statistical Analysis Fit to Model (time series) Advanced Data Analysis Clustering Meta analysis PCA Classification Promoter Analysis Survival analysis Regulatory Network Gene Ontology Gene Ontology (GO) is a collection of controlled vocabularies describing the biology of a gene product in any organism There are 3 independent sets of vocabularies, or ontologies: • Molecular Function (MF) – e.g. ”DNA binding” and ”catalytic activity” • Cellular Component (CC) – e.g. ”organelle membrane” and ”cytoskeleton” • Biological Process (BP) – e.g. ”DNA replication” and ”response to stimulus” Gene Ontology structure GO structure, example 2 KEGG pathways • KEGG PATHWAYS: – collection of manually drawn pathway maps representing our knowledge on the molecular interaction and reaction networks, for a large selection of organisms • 1. Metabolism – Carbohydrate, Energy, Lipid, Nucleotide, Amino acid, Other amino acid, Glycan, PK/NRP, Cofactor/vitamin, Secondary metabolite, Xenobiotics • • • • • 2. Genetic Information Processing 3. Environmental Information Processing 4. Cellular Processes 5. Human Diseases 6. Drug Development KEGG pathway example 1 KEGG pathway example 2 Cluster analysis and GO Analysis example: • Partitioning clustering of genes into e.g. 15 clusters based on expression profiles • Assignment of GO terms to genes in clusters • Looking for GO terms overrepresented in clusters Hypergeometric test • The hypergeometric distribution arises from sampling from a fixed population. 20 white balls out of 100 balls 10 balls • We want to calculate the probability for drawing 7 or more white balls out of 10 balls given the distribution of balls in the urn Yeast cell cycle Sampling Time series experiment: Y Y Y Y Y Y Y Time Gene expression profiles: Gene1 Gene2 Time R stuff Indexing of a matrix (used when you wish to select a subset of your data, e.g. specific rows or columns): • Example 1 rowindex <- 1:10 colindex <- 1:5 datamatrix[rowindex, colindex] # first 10 rows, first 5 columns datamatrix[1:10, 1:5] # gives the same as above “Missing” rowindex (or columnindex) means that all rows (or columns) are selected • Example 2 datamatrix[1:5,] # 5 first rows, all columns datamatrix[,5:10] # all rows, columns 5 to 10 datamatrix[,] # is the same as datamatrix