* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download supersecondar, tertiary and quaternary structure
Protein (nutrient) wikipedia , lookup
Self-assembling peptide wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein moonlighting wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Western blot wikipedia , lookup
Two-hybrid screening wikipedia , lookup
List of types of proteins wikipedia , lookup
Biochemistry wikipedia , lookup
Interactome wikipedia , lookup
Circular dichroism wikipedia , lookup
Homology modeling wikipedia , lookup
Protein adsorption wikipedia , lookup
Protein folding wikipedia , lookup
Protein domain wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
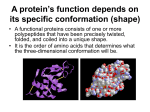
+ Super secondary structures + Super secondary structures motifs motifs or folds, are particularly stable arrangements of several elements of the secondary structure. Super secondary structures are usually produced by packing side chains from adjacent secondary structural elements close to each other. + Rules for secondary structure. Hydrophobic side groups must be buried inside the folds, therefore, layers must be created (b-a-b; a-a). a-helix and b-sheet, if occur together, are found in different structural layers. Adjacent polypeptide segments are stacked together. The b-sheet is the most stable. Motif + • Secondary structure composition, e.g. all a, all b, segregated a+b, mixed a/b • Motif = small, specific combinations of secondary structure elements, e.g. b-a-b loop + Super secondary Structures (Motifs) + + Tertiary protein structure Secondary structures fold and pack together to form tertiary structure Usually globular shape Tertiary structure stabilized by bonds between R groups (i.e. side chains) Intracellular protein tertiary structures mostly held together by weak forces. Extracellular tertiary structures stabilized by disulfide (covalent) bonds. + Three-dimensional structure of proteins Function of the protein depends on its structure. Each protein has a unique or nearly unique structure. Non-covalent interactions are the most important forces stabilizing the three dimensional structure of the protein. + + Interactions stabilizing tertiary structure : 1.Disulfide bonds: These strong, covalent bonds help stabilize the structure of proteins, and prevent them from becoming denatured in the extracellular environment. 2.Hydrophobic interactions 3.Hydrogen bonds 4. Ionic interactions + Tertiary structure - disulfide bond Covalent bond between sulfur atoms on two cysteine amino acids + Tertiary structure - H bond H Hydrogen bond bonds weak allowing to be broken and reformed easily Allows structural change produces ‘functional’ molecules • Ions on R groups form salt bridges through ionic bonds + Tertiary structure - hydrophobic forces Close attraction of non- polar R groups through dispersion forces Very weak but collective interactions over large area stabilize structure Repel polar and charged molecules/particles Tertiary Structure + 19 Tertiary Structure The interactions of the R groups give a protein its specific threedimensional tertiary structure. + Tertiary Structure • non-linear • 3 dimensional • global but restricted to the amino acid polymer • formed and stabilized by hydrogen bonding, covalent (e.g. disulfide) bonding, hydrophobic packing toward core and hydrophilic exposure to solvent • A globular amino acid polymer folded and compacted is somewhat functional (catalytic) and energetically favorable interaction! + Quaternary Structure of Proteins Many proteins consist of a single polypeptide chain, and are defined as monomeric proteins. others may consist of two or more polypeptide chains that may be structurally identical or totally unrelated. (Dimeric) The arrangement of these polypeptide subunits is called the quaternary structure of the protein. + Quaternary Structure of Proteins The biological function of some molecules is determined by multiple polypeptide chains – multimeric proteins Two kinds of quaternary structures: both are multi-subunit proteins. Homotypic: association between identical polypeptide chains. Heterotypic: interactions between subunits of very different structures. 23 The interactions within multi subunits are the same as that found in tertiary and secondary structures + Quaternary Structure • This structure for proteins that have more than one polypeptide chains. • It is the arrangement of protein subunits (protein that has more than one polypeptide chain) in three dimensional complex. • The interaction between subunits are stabilized by: • hydrogen bonds • electrostatic bonds • hydrophobic bonds e.g. of proteins having quaternary structure: • Hemoglobin (4 subunits) + 28 Summary of Structural Levels