* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Make:targeting signals
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Biochemistry wikipedia , lookup
Gene expression wikipedia , lookup
Magnesium transporter wikipedia , lookup
Protein (nutrient) wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Protein moonlighting wikipedia , lookup
List of types of proteins wikipedia , lookup
Signal transduction wikipedia , lookup
Ligand binding assay wikipedia , lookup
Metalloprotein wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Western blot wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Circular dichroism wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein adsorption wikipedia , lookup
PBio/NeuBehav 550: Biophysics of Ca2+ signaling
Week 4 (05/18/15)
Genetically expressible probes and FRET
Objectives for today:
• Why targeted and expressible probes
• Aequorin & GFP mixed with theory
• FRET Theory and photochemistry
• The first cameleons and GcAMPs
• Discuss a GCaMP paper
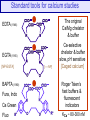
Standard tools for calcium studies
The original
Ca/Mg chelator
& buffer
EDTA (1946)
EGTA (1955)
[NP-EGTA]
[–—NP]
Ca-selective
chelator & buffer
slow, pH sensitive
[Caged calcium]
Ca Green
Roger Tsien’s
fast buffers &
fluorescent
indicators
Fluo
KCa ~ 80-300 nM
BAPTA (1980)
Fura, Indo
Advantages of proteins as indicators
Highly evolved binding sites
Can be further engineered by mutation
Sophisticated optical properties
Expressed by transfection, infection, transgenic; made within
cells; no loading; do not leak
Targetable:
by promoters to specific cell types at specific times in
organisms
by localization sequence's: to subcellular locations and
organelles in cells
Sequnce targeting of fluorescent constructs
Targeted to:
cytoplasm
N
fluorescent protein
C
ER
CRsig
fluorescent protein
KDEL
tpA
fluorescent protein
secretory
granules
nucleus
mitochondria
fluorescent protein
COX8
nls
fluorescent protein
Abbreviations:
CRsig = calreticulin signal sequence
KDEL = ER retention signal
tpA
= tissue plaminogen activator (a secreted protein)
nls
= nuclear localization signal
COX8 = cytochrome oxidase N-terminus
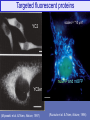
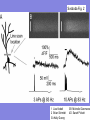
Targeted fluorescent proteins
YC2
scales = "10 mm"
nuGFP and mtBFP
YC3er
(Miyawaki et al. & Tsien, Nature, 1997)
(Ruzzuto et al. & Tsien, Nature, 1996)
Fluorescent proteins make Aequorea glow at 508 nm
The Nobel Prize in Chemistry 2008. Osamu Shimomura, Martin Chalfie, Roger Y. Tsien
Green fluorescent ring
---Shimomura O, Johnson FH, Saiga Y,
1962, Extraction, purification and
properties of Aequorin, a bioluminescent protein from the luminous
hydromedusan, Aequorea. J. Cell. Comp.
Physiol., 59: 223-239. [470 nm]
Aequorea victoria from Puget Sound
in brightfield and false color
---R.Y. Tsien, 1998, The Green
Fluorescent Protein, Annual Review of
Biochemistry 67, pp 509-544. [508 nm]
Aequorin: a bioluminescent
Ca2+ binding protein complex
containing coelenterazine
coelenterazine
M.W. = 22,514 with four E/F hands
Aequorin (Aeq) falls in the general heading of "luciferases" that bind a "luciferin" and
luminesce in response to a ligand. (The most famous of these is firefly luciferase
that can be used to measure ATP concentrations.)
Reaction:
Aeq + coelenterazine ----> Aeq.c [non-covalent complex]
Aeq.c + ~3 Ca2+
Ca3.Aeq.c*
----> Ca3.Aeq.c* + CO2
-----> Ca3.Aeq.c** + [blue photon--470 nm]
Aequorin is therefore a one-shot calcium detector with a non-linear Ca2+
dependence of luminescence. It is "consumed" by a detection event.
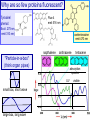
Why are so few proteins fluorescent?
Tyrosine/
phenol:
Fluo-4
emit 516 nm
Excit. 275 nm,
emit 310 nm)
coelenterazine
emit 470 nm
napthalene
anthracene
"Particle-in-a-box"
(think organ pipes)
absorption
spectra
UV
small box, short wave
large box, long wave
tetracene
visible
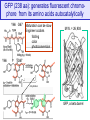
GFP (238 aa): generates fluorescent chromophore from its amino acids autocatalytically
Y66 G67
Y66
Maturation can be slow
Engineer codons
folding
color
photoconversion
M.W. = 26,938
G67
dehydration
N
C
GFP, a beta barrel
Engineering color in GFPs
Excitation spectra
5
4
5
400
500
Absorbance
Fluorescence intensity
4
Emission spectra
300
400
500
wavelength (nm)
600
600
700
wavelength (nm)
Roger Tsien's lab made a range of GFP-derived proteins of different colors by
mutation of the expression vector.
Absorption and fluorescence spectra reflect
internal energy levels
S1
S1
S0
S0
ground state
Absorption
wavelength
Em wl
Jablonski diagram
Absorber has several electronic states (S0, S1, S2, etc.). It also has vibrational
states whose close spacing means that photons of a range of close energies
can be absorbed. If the absorption spectrum has a second peak (at shorter
wavelength), it is for excitation to S2 or because the dye has several molecular
forms/conformations.
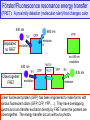
Förster/Fluorescence resonance energy transfer
(FRET): A proximity detector (molecular ruler) that changes color
440 nm
480 nm
hn
emission
hn
Separated: excitation
no FRET
440 nm
Close together:
FRET
YFP
CFP
hn
excitation
no 440 nm
excitation
no hn
CFP
FRET!
YFP
hn
535 nm
emission
Green fluorescent protein (GFP) has been engineered to make forms with
various fluorescent colors (GFP, CFP, YFP, …). They have overlapping
spectra and can transfer excitation directly by FRET when the proteins are
close together. The energy transfer occurs without a photon.
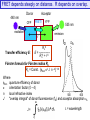
FRET depends steeply on distance. R depends on overlap.
Donor
Acceptor
440 nm
CFP
FRET!
YFP
excitation
r
Transfer efficiency E:
535 nm
emission
R o6
E = ------------R o6 + r 6
fD
eA
Förster formula for Förster radius Ro
Ro = Const. {fdon k2 J n –4} 1/6
Where
fdon quantum efficiency of donor
k
orientation factor (0 – 4)
n
local refractive index
500
600
J
"overlap integral" of donor fluorescence (fD) and acceptor absorption eA
J=
l = wavelength
FRET as a ‘Spectroscopic Ruler’
The efficiency of energy transfer is proportional to the inverse of
the sixth power of the distance separating the donor and
acceptor fluorophore
ECFP/EYFP
Förster distance 30 Å
Förster distance 50 Å
e.g., ECFP/EYFP
Förster distance 70 Å
E % decreases with
the distance between
donor and acceptor
Two fluorophores separated by Förster distance (r = Ro) have E transfer of 50%
More steps in the Jablonski diagram
internal
conversion
(1 ps)
(polar)
solvent
relaxation
(100 ps)
competition
for re-radiation,
quench, FRET,
or other nonradiative (3 ns)
absorption
(1 fs)
knr
fluorescence
Donor
hnFRET
quench
FRET
Acceptor
FRET speeds donor F and slows acceptor F
competition
for re-radiation,
quench, FRET
(polar) solvent
relaxation
(100 ps)
internal
conversion
(1 ps)
absorption
(1 fs)
emission intensity
Donor
knr
fluorescence
CFP
hnFRET
quench
Acceptor
FRET
YFP
Ca2+-bound CaMeleon
530 nm from
EYFP by FRET
480 nm from
ECFP
0
2
time (ns)
4
Fluorescence
lifetime imaging is a
6 way to image FRET
Fluorescence decays recorded with YC3.1 cameleon dissolved in buffer.
Excitation at 420 nm excites the ECFP part. (Habuchi et al. Biophys J, 2002)
A family of Ca2+-sensitive switches and buffers
helix-loophelix makes
E-F hand
x
x
x
x
Calmodulin
MW ~ 17 kDa
KCa ~ 14 mM
for free
calmodulin
Calmodulin (CaM) : An abundant 149 amino acid, highly conserved cytoplasmic protein with 4 binding sites for Ca2+ each formed by "EF-hands."
Many other homologous Ca2+ binding proteins of this large EF-hand family
act as Ca switches and Ca buffers. The Ca2+ ions bind cooperatively and
become encircled by oxygen dipoles and negative charge. CaM complexes with many proteins, imparting Ca2+-dependence to their activities.
Calmodulin folds around a target helix
M13: MLCK
peptide
4 Ca
CaM
Binding of Ca2+ to CaM causes CaM
to change conformation. Binding of
CaM to targets can increase the Ca2+
binding affinity of CaM greatly.
The target peptide in this crystal structure is the regulatory domain of
smooth-muscle myosin light-chain kinase (MLCK). The interaction of
CaM and MLCK allows smooth muscle contraction to be activated in a
Ca2+-dependent manner. (Meador WE, Means AR & Quiocho, 1992.)
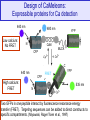
Design of CaMeleons:
Expressible proteins for Ca detection
440 nm
Low calcium:
No FRET
480 nm
YFP
C
N
CaM
MLCK
CFP
C
440 nm
CFP
High calcium:
FRET
N
FRET
YFP
535 nm
Two GFPs in one peptide interact by fluorescence resonance energy
transfer (FRET). Targeting sequences can be added to direct constructs to
specific compartments. (Miyawaki, Roger Tsien et al., 1997)
Ca-sensitive cameleon emission spectra
Note two peaks
no Ca
emission intensity
Ca
YC3.1
cameleon
more
FRET
Emission wavelength (nm)
(Miyawaki, Roger Tsien et al., 1997)
Ca-sensitive FRET reporter. How do calciums bind?
(Miyawaki et al., 1997)
green cameleon 1 fluorescence ratios
510/445 nm emission ratio
1.0
E104
C
lower
affinity
E31 N
higher affinity
GC1
GC1/E31Q
GC1/E104Q
free calcium (M)
Calcium binding and the conformation change can be tailored by making
mutations in the EF hand regions of the calmodulin. Glutamate E31 is in the
first EF hand (at p12') and E104 is in the third EF hand (also at p12').
ER-directed Cameleon sees stores
SERCA inhibition
PLC activation
Caex
PC12 cells are transfected with D1-ER, a Roger Tsien cameleon directed to the
ER. SERCA pump blocker BHQ shows efflux,. ATP makes IP3 production and
calcium release from ER. (Dickson,....,Hille, 2012)
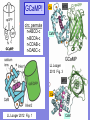
GCaMP!
circ. permute
n-ABCD-c
n-BCDA-c
n-CDAB-c
n-DABC-c
Ca
M13
CaM
LL Looger
2013 Fig. 3
M13
Ca
LL Looger 2012 Fig. 1
CaM
GCaMP descendants
RCaMP1h
First generation (2001) was slow, green, and pH dependent.
Keeps getting better, brighter, faster, more colors.
LL Looger Front Mol Neurosci. 2013
Mao T, O'Connor DH, Scheuss V, Nakai J, Svoboda K.
Characterization and subcellular targeting of GCaMPtype genetically-encoded calcium indicators. PLoS One.
2008 Mar 19;3(3):e1796.
Each figure will be described by a student--as if you are
teaching it to us for the first time.
Further questions will come from the audience.
--5 min per fig--one panel at a time
--give it a title
--explain axes and subject
--ask leading questions to get students
to discuss--what is being tested and
what is concluded?
1 Liza Kobelt
2 Brian Schmidt
3A Kelly Duong
3B Michelle Gianmarco
4D Sarah Pickett
Svoboda Fig. 1
1 Liza Kobelt
2 Brian Schmidt
3A Kelly Duong
3B Michelle Gianmarco
4D Sarah Pickett
Svoboda Fig. 2
1 Liza Kobelt
2 Brian Schmidt
3A Kelly Duong
3B Michelle Gianmarco
4D Sarah Pickett
Svoboda Fig. 3A
1 Liza Kobelt
2 Brian Schmidt
3A Kelly Duong
3B Michelle Gianmarco
4D Sarah Pickett
Svoboda Fig. 3B
1 Liza Kobelt
2 Brian Schmidt
3A Kelly Duong
3B Michelle Gianmarco
4D Sarah Pickett
Svoboda Fig. 4D
1 Liza Kobelt
2 Brian Schmidt
3A Kelly Duong
3B Michelle Gianmarco
4D Sarah Pickett