* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Regulation of Gene Expression
Genomic imprinting wikipedia , lookup
Magnesium transporter wikipedia , lookup
Ridge (biology) wikipedia , lookup
Non-coding DNA wikipedia , lookup
Community fingerprinting wikipedia , lookup
RNA silencing wikipedia , lookup
Molecular evolution wikipedia , lookup
Gene desert wikipedia , lookup
RNA interference wikipedia , lookup
Genome evolution wikipedia , lookup
Transcription factor wikipedia , lookup
Protein moonlighting wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Histone acetylation and deacetylation wikipedia , lookup
List of types of proteins wikipedia , lookup
Polyadenylation wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Non-coding RNA wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Expression vector wikipedia , lookup
Messenger RNA wikipedia , lookup
Gene expression profiling wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epitranscriptome wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Gene regulatory network wikipedia , lookup
Gene expression wikipedia , lookup
Regulation of Gene
Expression
{
Chapter 13
Regulation of gene expression in eukaryotes:
Levels of control of gene expression
Short term control
(to meet the daily needs of the organism)
Long term control
(gene regulation in development/differentiation)
Differences between prokaryotes and eukaryotes:
•
Prokaryote gene expression typically is regulated by an operon, the
collection of controlling sites adjacent to polycistronic proteincoding sequences.
•
Eukaryotic genes also are regulated in units of protein-coding
sequences and adjacent controlling sites, but operons are not
known to occur.
•
Eukaryotic gene regulation is more complex because eukaryotes
possess a nucleus.
(transcription and translation are not coupled).
•
Two “categories” of eukaryotic gene regulation exist:
Short-term - genes are quickly turned on or off in response to the
environment and demands of the cell.
Long-term - genes for development and differentiation.
An operon typically includes:
Regulator gene- this codes for a DNA-binding protein that
acts as a repressor
Promoter – DNA sequence that binds RNA polymerase
Operator- portion of DNA where an active repressor binds
Structural Genes- codes for enzymes and proteins needed for
the operons metabolic pathway
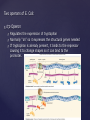
Two operons of E. Coli:
trp Operon
Regulates the expression of tryptophan
Normally “on” so it expresses the structural genes needed
If tryptophan is already present, it binds to the repressor
causing it to change shapes so it can bind to the
promoter.
lac Operon
Regulation for lactose metabolism
The repressor is normally bound to the operator to turn “off”
gene expression
In the presence of lactose, lactose binds to the repressor
causing it to change shape so DNA polymerase can easily
bind to the promotor.
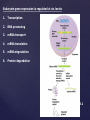
Eukaryote gene expression is regulated at six levels:
1.
Transcription
2.
RNA processing
3.
mRNA transport
4.
mRNA translation
5.
mRNA degradation
6.
Protein degradation
Fig. 18.1
1. Transcription control of gene regulation is controlled by:
1.
Promoters
•
Occur upstream of the transcription start site.
•
Some determine where transcription begins (e.g., TATA),
whereas others determine if transcription begins.
•
Promoters are activated by specialized transcription factor (TF)
proteins (specific TFs bind specific promoters).
•
One or many promoters (each with specific TF proteins) may
occur for any given gene.
•
Promoters may be positively or negatively regulated.
1. Transcription control of gene regulation is controlled by:
2.
Enhancers
•
Occur upstream or downstream of the transcription start site.
•
Regulatory proteins bind specific enhancer sequences; binding
is determined by the DNA sequence.
•
Loops may form in DNA bound to TFs and make contact with
upstream enhancer elements.
•
Interactions of regulatory proteins determine if transcription
is activated or repressed (positively or negatively regulated).
More about promoters and enhancers:
•
Some regulatory proteins are common in all cell types, others are
specific.
•
Each promoter and enhancer possesses a specific set of proteins
(coactivators) that determines expression.
•
Rate of gene expression is controlled by interaction between
positive and negative regulatory proteins.
•
Combinatorial gene regulation; enhancers and promoters bind many
of the same regulatory proteins, implying lots of interaction with
fine and coarse levels of control.
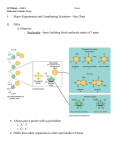
Short-term - transcriptional control of galactose-utilizing genes in yeast:
•
3 genes (GAL1, GAL7, & GAL 10) code enzymes that function in the
galactose metabolic pathway.
•
GAL1
galactokinase
•
GAL7
galactose transferase
•
GAL10
galactose epimerase
•
Pathway produces d-glucose 6-phosphate, which enters the
glycolytic pathway and is metabolized by genes that are
continuously transcribed.
•
In absence of galactose, GAL genes are not transcribed.
•
GAL genes rapidly induced by galactose and absence of glucose.
•
Analagous to E. coli lac operon repression by glucose.
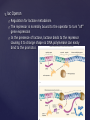
Galactose metabolizing pathway of yeast.
Short-term - transcriptional control of galactose-utilizing genes in yeast:
•
GAL genes are near each other but do not constitute an operon.
•
Additional unlinked gene, GAL4, codes a repressor protein that binds
a promoter element called an upstream activator sequence (UASG).
•
UASG is located between GAL1 and GAL10.
•
Transcription occurs in both directions from UASG.
•
When galactose is absent, the GAL4 product (GAL4p) and another
product (GAL80p) bind the UASG sequence; transcription does not
occur.
•
When galactose is added, a galactose metabolite binds GAL80p and
GAL4p amino acids are phosphorylated.
•
Galactose acts as an inducer by causing a conformation change in
GAL4p/GAL80p.
Activation model of GAL genes in yeast.
2. RNA processing control:
•
RNA processing regulates mRNA production from precursor RNAs.
•
Two independent regulatory mechanisms occur:
•
Alternative polyadenylation
= where the polyA tail is added
•
Alternative splicing
= which exons are spliced
•
Alternative polyadenylation and splicing can occur together.
•
Examples:
•
Human calcitonin (CALC) gene in thyroid and neuronal cells
•
Sex determination in Drosophila
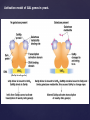
Alternative polyadenylation and splicing of the human CACL gene in
thyroid and neuronal cells.
Calcitonin
gene-related
peptide
Alternative splicing in
sex determination of
Drosophila
•Sex is determined by
X:A ratio.
•Sxl (sex lethal) gene
determines the
pathways for males
and females.
•If X:A = 1, all introns
and exon 3 (which
contains the stop
codon) are removed.
•If X:A = 0.5, no
functional protein is
produced.
3. mRNA transport control:
•
Eukaryote mRNA transport is regulated.
•
Some experiments show ~1/2 of primary transcripts never leave the
nucleus and are degraded.
•
Mature mRNAs exit through the nuclear pores.
4. mRNA translation control:
•
Unfertilized eggs are an example, in which mRNAs (stored in the
egg/no new mRNA synthesis) show increased translation after
fertilization).
•
Presence or absence of the 5’ cap and the length of the poly-A tail at
the 3’ end can determine whether translation takes place and how
long the mRNA is active.
•
Conditions that affect the length of the poly-A tail or leads to the
removal of the cap may trigger the destruction of an mRNA.
5. mRNA degradation control:
•
All RNAs in the cytoplasm are subject to degradation.
•
tRNAs and rRNAs usually are very stable; mRNAs vary considerably
(minutes to months).
•
Stability may change in response to regulatory signals and is
thought to be a major regulatory control point.
6. Post-translational control - protein degradation:
•
Proteins can be short-lived (e.g., steroid receptors) or long-lived
(e.g., lens proteins in your eyes).
•
This is a cells last change to affect gene expression.
•
Protease, enzymes that break down proteins, are confined to
lysosomes and proteasomes.
•
When a protein is tagged by a signaling protein, it enters a
proteasome to be degraded.
Summary and contrasts:
Prokaryotes control expression by:
Transcription
Eukaryotes control expression by:
Transcription
RNA processing
mRNA transport
mRNA translation
mRNA degradation
Protein degradation
Fig. 18.1