* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Beta sheets are twisted
Expression vector wikipedia , lookup
Gene expression wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Catalytic triad wikipedia , lookup
Interactome wikipedia , lookup
Point mutation wikipedia , lookup
G protein–coupled receptor wikipedia , lookup
Magnesium transporter wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Evolution of metal ions in biological systems wikipedia , lookup
Genetic code wikipedia , lookup
Structural alignment wikipedia , lookup
Biosynthesis wikipedia , lookup
Western blot wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Zinc finger nuclease wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Biochemistry wikipedia , lookup
Homology modeling wikipedia , lookup
Anthrax toxin wikipedia , lookup
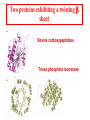
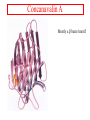
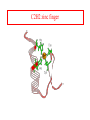
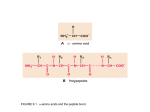
Protein Structure Lecture 2/26/2003 beta sheets are twisted • Parallel sheets are less twisted than antiparallel and are always buried. • In contrast, antiparallel sheets can withstand greater distortions (twisting and betabulges) and greater exposure to solvent. The twist is due to chiral (l)- amino acids in the extended plane. This chirality gives the twist and distorts Hbonding. A tug of war exists between conformational energies of the side chain and maximal Hbonding. Two proteins exhibiting a twisting b sheet Bovine carboxypeptidase Triose phosphate isomerase Connections between adjacent b sheets Sheet facts •Repeat distance is 7.0 Å •R group on the Amino acids alternate up-down-up above and below the plane of the sheet •2 - 15 amino acids residues long •2 - 15 strands per sheet •Ave of 6 strands with a width of 25 Å •parallel less stable than anti-parallel •Anti-parallel needs a hairpin turn •Tandem parallel needs crossover connection which has a right handed sense Non-repetitive regions Turns - coils or loops link regions of secondary structure 50% of structure of globular proteins are not repeating structures b bends type I and type II :hairpin turn between anti parallel sheets Reverse Turns Type I f2 = -60o, y2 = -30o f3 = -90o, y3 = 0o Type II f2 = -60o, y2 = 120o f3 = 90o, y3 = 0o two-residue turns Protein Structure Terminology Folding motifs (super secondary structure) Certain amino sequences have patterns to their folding. A. bab motif, B. b hairpin C. aa motif beta-alpha-beta • parallel beta-strands connected by longer regions containing alpha-helical segments • almost always has a right-handed fold Helix-turn-helix • • • • • loop regions connecting alpha-helical segments can have important functions e.g. EF-hand and DNA-binding EF hand loop ~ 12 residues polar and hydrophobic a.a. conserved positions Glycine is invariant at the sixth position The calcium ion is octahedrally coordinated by carboxyl side chains, main chain groups and bound solvent Protein Folds There is an estimate of about 10000 different folding patterns in proteins About half of the proteins fall into a few dozen folding patterns. Those proteins related by structure are called families. A large Family are the c cytochromes (see Figure 6-31 pg 147 in FOB.) The b barrel has several types of structures that have been mimicked in art. A. rubredoxin B. Human prealbumin or porins C. Triose phosphate isomerase Concanavalin A Mostly a b barrel motif Carbonic anhydrase H2CO3 - CO2 + H2O Nucleotide binding-Rossmann Fold Glyceraldehyde-3phosphate dehydrogenase Binding NADH in the Rossmann fold. Zinc fingers C2H2 zinc finger: It is characterized by the sequence CX2-4C....HX2-4H, where C = cysteine, H = histidine, X = any amino acid. C4 zinc finger: Its consensus sequence is CX2CX13CX2CX1415CX5CX9CX2C. The first four cysteine residues bind to a zinc ion and the last four cysteine residues bind to another zinc ion C6 zinc finger. It has the consensus sequence CX2CX6CX56CX2CX6C. The yeast's Gal4 contains such a motif where six cysteine residues interact with two zinc ions C2H2 zinc finger Zinc Finger DNA-binding Summary Chapter 6 • Four levels of protein structure – – – – • • • • • Primary Secondary Tertiary Quaternary Peptide bond (w bond) Sheets and helices (f and y bonds) Tertiary structure (fibrous or globular) Structure determination and fold space Protein folding discussed after kinetics -lecture 19