* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Valyl tRNA-Synthestase - Illinois State University
Survey
Document related concepts
Catalytic triad wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Proteolysis wikipedia , lookup
Peptide synthesis wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Metalloprotein wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Protein structure prediction wikipedia , lookup
Epitranscriptome wikipedia , lookup
Genetic code wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Biochemistry wikipedia , lookup
Anthrax toxin wikipedia , lookup
Transcript
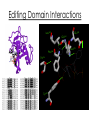
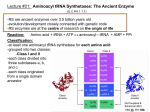
Overall Structure Editing Domain (“2nd Sieve”) 862 amino acids 25 α-helices 30 β-strands Catalytic Domain (“1st Sieve”) C-Terminal Coiled-Coil Domain Anticodon Binding Domain Function • Aminoacyl tRNA synthetases are enzymes that catalyze the esterification of a specific amino acid to a compatible cognate tRNA to form an aminoacyltRNA • Class I vs. Class II: – 2’-OH, then 3’-OH – Directly to 3’-OH “Double-Sieve” Concept Aminoacylation Site Interactions Hydrophobic Pocket Trp456 Trp491 Asp81 Pro42 Pro41 Ile491 Asn44 KMSKS Loop Met529 Editing Domain Interactions Leu269 Glu281 Glu261 Phe264 Thr214 Tyr337 Leu278 Editing Hydrophilic Pocket Anticodon Binding Domain *nucleotides are in green, amino acids are in yellow Kinetics for C-term. Domain o phosphates on A20 & A21 interact through salt bridges with Arg818 and Arg843 o G19 & C56 crucial of correct positioning of 3’ CCA end of tRNA into aminoacylation catalytic site References 1. 2. 3. 4. Fukai, S.; Nureki, O.; Sekine, S.; Shimada, A.; Tao, J.; Vassylyev, D.G.; Yokoyama, S. Structural Basis for Double-Sieve Discrimination of L-Valine from L-Isoleucine and LThreonine by the Complex of tRNAVal and Valyl-tRNA Synthetase. Cell 2000, 103, 793-803. Fukai, S.; Nureki, O.; Sekine, S.; Shimada, A.; Vassylyev, D.G.; Yogoyama, S. Mechanism of molecular interactions for tRNAVal recognition by valyl-tRNA synthetase. RNA 2003, 9, 100111. Fukunaga, R.; Yokoyama, S. Structural Basis for Non-cognate Amino Acid Discrimination by the Valyl-tRNA Synthetase Editing Domain. J. Bio. Chem. 2005, 280 (33), 29937-29945. Liu, M.; Chu, W.; Liu, J.C.H.; Horowitz, J. Role of acceptor stem conformation in tRNAVal recognition by its cognate synthetase. Nucleic Acids Res. 1997, 25, 4883-4890.