* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download CHAPTER 39: The Genetic Code
Survey
Document related concepts
Protein (nutrient) wikipedia , lookup
Molecular evolution wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Non-coding RNA wikipedia , lookup
Bottromycin wikipedia , lookup
Butyric acid wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Cell-penetrating peptide wikipedia , lookup
Citric acid cycle wikipedia , lookup
Peptide synthesis wikipedia , lookup
Epitranscriptome wikipedia , lookup
Point mutation wikipedia , lookup
Protein structure prediction wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Transfer RNA wikipedia , lookup
Biochemistry wikipedia , lookup
Transcript
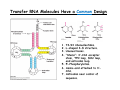
CHAPTER 39: The Genetic Code Problems 1,3,5,7-9,12,13,15,18-19,21,23,25-26 39:1 The Genetic Code Links Nucleic Acid and Protein Information 1. Three nucleotides encode an amino acid. 2. The code is nonoverlapping. 3. The code has “no” punctuation. 4. The code has directionality. 5. The code is degenerate. Three nucleotides encode an amino acid. •DNA – only four bases (A,T,G,C) •Must code for 20 amino acids •Two-base code: 42 = 16 combinations •Four-base code: 44 = 256 combinations •Three-base code: 43 = 64 combinations The code is nonoverlapping The code has “no” punctuation. The code has directionality and is degenerate. (5’-UUU-3’) (5’-UUC-3’) The Genetic Code is Nearly Universal Transfer RNA Molecules Have a Common Design 1. 2. 3. 4. 73-93 ribonucleotides. L-shaped 3-D structure. Unusual bases. “Stems”: 3’-CCA acceptor stem, TΨC loop, DHU loop, and anticodon loop. 5. 5’-Phosphorylation. 6. Amino acid attached to 3’CCA. 7. Anticodon near center of sequence. Some Transfer RNA Molecules Recognize More Than One Codon Because of Wobble in BasePairing • tRNA molecules are named for the amino acid that they carry (e.g. tRNAPhe) • Base pairing between codon and anticodon is governed by rules of Watson-Crick (AU, G-C) • However, the 5’ anticodon position has some flexibility in base pairing (the “wobble” position) 3’ 5’ (5’) (3’) tRNAPhe AAG 5’ UUC mRNA (Phe) Wobble position 3’ Variable position The Synthesis of Long Proteins Requires a Low Error Frequency 39:2 Amino Acids Are Activated by Attachment to Transfer RNA Amino Acid + ATP aminoacyl-AMP + tRNA PPi + H2O Amino Acid + tRNA + H2O aminoacyl-AMP + PPi aminoacyl-tRNA + AMP 2 Pi aminoacyl-tRNA + AMP + 2Pi Synthetases Have Highly Discriminating Amino Acid Activation Sites • Attachment of the correct amino acid to the corresponding tRNA is a critical step • Synthetase binds ATP and the correct amino acid (based on size, charge, hydrophobicity) • Synthetase then selectively binds specific tRNA molecule based on structural features • Synthetase may recognize the anticodon as well as the acceptor stem Val-tRNAThr does not happen. However, Ser-tRNAThr can occur 1/100 or 1/1000 times. Proofreading by Aminoacyl-tRNA Synthetases Too Small Too Large Aminoacyl-tRNA Synthetases Recognize Anticodon Loop and Acceptor Stem 39:3 A Ribosome is a Ribonucleoprotein DNA Replication: 5’→3’ Transcription: 5’→3’ Translation: 5’→3’