* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download You are what you eat? Plant nutrient status and the
Plant breeding wikipedia , lookup
Genetic engineering wikipedia , lookup
Genomic imprinting wikipedia , lookup
Western blot wikipedia , lookup
Magnesium transporter wikipedia , lookup
Non-coding DNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Proteolysis wikipedia , lookup
Interactome wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Gene nomenclature wikipedia , lookup
Community fingerprinting wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Protein–protein interaction wikipedia , lookup
Gene regulatory network wikipedia , lookup
Expression vector wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Gene expression profiling wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Gene expression wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Point mutation wikipedia , lookup
Molecular evolution wikipedia , lookup
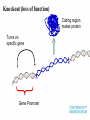
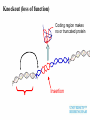
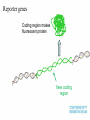
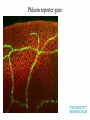
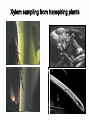
Plant adaptation to changing environments: A role for GM Dr Jeremy Pritchard @DrJPritchard Ecology Organisms Cells Molecules Molecules: Transcription and Translation DNA mRNA Protein Population growth Climate change Conventional breeding has been going on for 10,000 years Why Sequence Genomes? o Will eventually tell us what genes do – Leads to Medical Applications – Leads to Agricultural Applications o Tells us about evolution o Tell us how we develop o Tell us how we are different to cabbages, mice and chimps Applications of GM crops Examples from my research o Knockouts o Localisation o Transcriptomics What does a gene do? Where is it expressed? How do genes respond? Must combine Molecular with Physiology Knockout (loss of function) Coding region makes protein Turns on specific gene Gene Promoter Knockout (loss of function) Coding region makes no or truncated protein Insertion Reporter genes Remove coding region Reporter genes Coding region makes fluorescent protein New coding region Phloem reporter gene Salinity – the silent flood Salt crosses membranes through protein ‘gates’ These gates control salt levels in xylem Salt Salt Bioinformatics- Arabidopsis – the model plant ctgtcttctcactaaactccaaaacccaccggaaaaatgatta cgtggcacgacttgtacaccgtcctcaccgccgtggtaccact ttacgtagctatgattctttacggcaatatttcacgcctacggat ccgtacagtggtggaagatattctcaccagaccagtgctccg gcacaaccgcttcgtcgctatcttcgccgtccctctcctctccttc cacttcatctccaccaacgat CHx21 DNA Sequence MSSGAPLNVTNPNYDIEESRFGKIVCYDQSLLF EKREQKGWESGSTLASSLPFFITQLFVANLSYR VLYYLTRPLYLPPFVAQILCGLLFSPSVLGNTRFI IAHVFPYRFTMVLETFANLALVYN Amino acid Sequence Computer says: it looks like a salt transporter Immunology- protein is detected in root endodermis 10µm Is the computer right? Find out; make knockout mutants ctgtcttctc accgtcctca atcacgccta tcaaccgctt acgatcctta actaaactcc ccgccgtggt cggatccgta cgtcgctatc cgccatgaat aaaacccacc accactttac cagtggtgga ttcgccgtcc ggaaaaatga gtagctatga agatattctc ctctcctctc ttacgtggca ttct ttacg accagaccag cttccacttc cgacttgtac tggcaatatt tgctccggca atctccacca Insert extra sequence: This means stop – no protein is made Xylem sampling from transpiring plants Leaf salt is lower in mutant xylem and leaf 100 Wild type Mutant Leaf salt 75 50 25 0 control saline 5 Wild type Salt in xylem 4 mutant 3 2 1 0 Control Saline Bioinformatics • 5 similar sequences to CHX21 • In pairs on chromosome 1 & 2 1. Duplication 2. Translocation 3. Diversification CHX5 CHX14 CHX23 Chromosome 1 CHX5 CHX8 CHX8’ CHX14 CHX13 CHX13’ CHX23 CHX21 CHX21’ Chromosome 2 Evolution in action From this…………... To this…………... ……………….a better adapted plant Dr Jeremy Pritchard @DrJPritchard Slides and other resources are available as PowerPoint Email me: [email protected] or go to http://www.birmingham.ac.uk/schools/bi osciences/outreach