* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download A Comparative Genomic Method for Computational
Metalloprotein wikipedia , lookup
Lipid signaling wikipedia , lookup
Paracrine signalling wikipedia , lookup
Biochemical cascade wikipedia , lookup
Magnesium transporter wikipedia , lookup
Western blot wikipedia , lookup
Clinical neurochemistry wikipedia , lookup
Biochemistry wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Fatty acid synthesis wikipedia , lookup
Expression vector wikipedia , lookup
Signal transduction wikipedia , lookup
Gene regulatory network wikipedia , lookup
Transcription factor wikipedia , lookup
Non-coding DNA wikipedia , lookup
Gene expression wikipedia , lookup
Fatty acid metabolism wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Genomic library wikipedia , lookup
Point mutation wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Biosynthesis wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Two-hybrid screening wikipedia , lookup
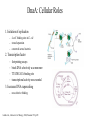
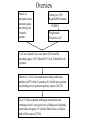
Comparative genomic analysis of lipid biosynthesis and metabolism components of the DnaA regulon Megon Walker Simon Kasif Introduction DnaA: Cellular Roles 1. Initiation of replication – 4 oriC binding sites in E. coli – strand separation – conserved across bacteria 2. Transcription factor – – – – footprinting assays binds DNA selectively as a monomer TTATNCACA binding site transcriptional activity non-essential 3. Increases DNA supercoiling – non-selective binding Lodish et al., Molecular Cell Biology, 2000, Freeman: NY. p 459. DnaA: Lipid biosynthesis • Temperature sensitive dnaA transcription factor prohibitive mutants at nonpermissive temperature – period of generation of cell growth NOT prolonged – altered lipid synthesis protein levels • increased beta-ketoacyl synthase II (fadL) • increased long-chain fatty acid transport protein (fabF) – altered fatty acid composition of cell membrane phospholipids • phosphatidylethanolamine (PE) • phosphatidylglycerol (PG) • How does DnaA regulate genes controlling the phospholipid fatty acid composition and flagella formation? Suzuki, E. et al., Mol Microbiol, 1998. 28(1): p. 95-102. Ohba, A. et al., FEBS Lett, 1997. 404(2-3): p. 125-8. DnaA: Transcription Factor • Goal – DnaA regulon characterization via identification of genes with DnaA binding sites in promoter regions – Repressor (dnaA, rpoH, uvrB, mioC, fadL) – Activator (nrdAB, glpD, polA) • Obstacles – 9mers resembling binding site occur frequently in E. coli genome – not all sites matching consensus are actually bound by DnaA (ftsAQ) – some experimentally conserved binding sites differ from consensus – known DnaA regulated genes not functionally related (replication, lipid synthesis, house keeping genes) Messer, W. et al., Mol Microbiol, 1997. 24(1): p. 1-6. Comparative genomics • Haemophilus influenza genome completed in 1995, ~100 genomes sequenced since Availability of complete genomes of related bacteria allows comparative analysis of regulatory patterns (gene number, content, and order in groups of organisms) Conservation of candidate DnaA binding sites across species is additional evidence of regulatory functionality If a regulator is conserved in several genomes • • • – its regulon and binding sites in these genomes are conserved as well – – true sites occur upstream of orthologous genes false sites are scattered at random across the genome Methods Overview Datasets of transcription units, upstream regions, and orthology for 8 bacterial genomes Training set (12/9) RegulonDB & literature PATSER Weight matrix Threshold: μ-2σ2 E. coli sets of putative site scores above 6.0 bit cutoff in noncoding regions (1031 Watson/1051 Crick). Performed for all 8 genomes. Reference E. coli k12 transcription units sharing orthologous members with TUs from 2+ genomes, all of which have upstream DnaA binding sites in upstream regulatory regions (164/120) Sets of 3+ DnaA-regulated, orthologous transcription units containing at least 1 cross-species pair of binding sites displaying conservation of sequence (2+ identical DnaA boxes) or location (within 20 base pairs) (127/88) Three Ortholog Selection Criteria 1. Selection of genomes for comparative study – 2. 3. E. coli k12, H.influenzae, S. typhimurium Lt2, V. cholerae, P. aeruginosa, Y. pestis, B. subtilis, B. halodurans Transcription unit (TU) designation – open reading frames transcribed in the same direction – separated by less than 100 intergenic nucleotides Pairwise identification of orthologs to genes in reference genomes: – – – reciprocal pairwise TBLASTN searches between all annotated genes of reference E. coli k12 and the other 7 organisms bidirectional best matches lower similarity threshold 10-20 Altschul, S. et al., JMB,1990. 215: p 403-410. PATSER: Position Weight Matrix Construction Alignment matrix dnaA TTATCCACA mioC TTTTCCACA rpoH TTATTCACA TTATCCACA uvrB TTATCCACT TTATCCACA nrdA TTATCCACA TTATGCACT polA TTATCCACA dam TTCTCCACA guaB TTATACAGA fadL TTATACAAA Hertz, G. et al, Comput Appl Biosci, 1990. 6(2): p. 81-92. A C G T | | | | 0 0 0 12 0 0 0 12 10 1 0 1 0 0 0 12 2 8 1 1 0 12 0 0 12 0 0 0 G 1 10 1 0 10 0 0 2 A C T -2.56 -2.56 -2.56 1.16 -2.56 -2.56 -2.56 1.16 0.99 -0.80 -2.56 -1.09 -2.56 -2.56 -2.56 1.16 -0.51 1.12 -0.78 -1.09 -2.56 1.52 -2.56 -2.56 1.16 -2.56 -2.56 -2.56 -1.09 1.34 -0.78 -2.56 0.99 -2.56 -2.56 -0.52 Weight matrix Training Set Gene Site Sequence Position Score dnaA ttatccaca -211 10.59 mioC tttttcaca -302 6.31 rpoH ttattcaca -131 8.38 ttatccaca -107 10.59 ttatccact -419 9.09 ttatccaca -405 10.59 ttatccaca -162 10.59 ttatgcact -150 7.19 polA ttatccaca -132 10.59 dam ttctccaca -30 8.81 guaB ttatacaga -45 6.84 fadL ttatacaaa -27 6.53 uvrB nrdA http://www.bio.cam.ac.uk/cgi-bin/seqlogo/logo.cgi Salgado, H. et al., Nucleic Acids Res, 2001 Jan 1. 1(72-4). Results Putative dnaA regulon: functional classifications • Training Set (8/9) • Lipid synthesis (6) • Information Transfer: transcription, translation, DNA repair, ribosomal assembly, nucleotide synthesis (13) • Coenzyme Metabolism (4) • Carbohydrate Transport & Metabolism (5) • Amino Acid Transport & Metabolism (6) • Energy Production and Conservation (6) • Putative/hypothetical ORFs (42) Tatusov, R. et al.. Nucleic Acids Res, 2001. 29(1): p. 22-8. Riley, M. et al.. J Mol Biol, 1997. 268(5): p. 857-68. acyl carrier protein (acpP) acpP-fabF Site Sequence Position Score ecolik12 ttatacact -54 7.45 paer ttttccata -17 6.09 vchol tttttcaca -77 6.30 stlt2 ttatacact -54 7.55 ypes ttatacact -54 7.40 • AcpP is an acyl carrier protein involved in lipid biosynthesis • co-transcribed upstream of FabF (fatty acid transport protein) Cronan Jr, J.E. et al., Escherichia coli and Salmonella typhimurium: cellular and molecular biology, F. Neidhardt, et al., Editors.1996, American Society for Microbiology: Washington DC. p. 615. carboxylase transferase (accD) accD-folD-dedD Site Sequence Position Score ecolik12 ttatccaaa -119 8.17 vchol taatccaca -72 6.91 stlt2 ttatccaaa -113 8.26 • accD transcribes a subunit of carboxylase transferase • performs the initial step of fatty acid synthesis Cronan Jr, J.E. et al., Escherichia coli and Salmonella typhimurium: cellular and molecular biology, F. Neidhardt, et al., Editors.1996, American Society for Microbiology: Washington DC. p. 614. PlsC (plsC) plsC-sufI Site Sequence Position Score ecolik12 ttttccaga -77 6.40 hinf ttatgcaga -162 6.40 stlt2 ttttccaga -78 6.44 Cronan Jr, J.E. et al., Escherichia coli and Salmonella typhimurium: cellular and molecular biology, F. Neidhardt, et al., Editors.1996, American Society for Microbiology: Washington DC. p. 619. • PlsC is involved in phospholipid biosynthesis preceding formation of cell membrane lipids PE and PG acyl carrier protein phosphodiesterase (acpD) acpD Site Sequence Position Score ecolik12 ttattcaca -54 8.38 bhal ttatgcaaa -379 6.09 paer ttataaaca -106 7.11 stlt2 ttatccgca -424 6.91 stlt2 ttttccaga -345 6.44 stlt2 ttattcaca -62 8.48 ypes ttatgcaga -60 6.52 ypes ttatccact -444 9.10 • AcpD is classified as an acyl carrier protein phosphodiesterase • highest scoring putative DnaA binding site Psd (psd) yjeQ-psd Site Sequence Position Score ecolik12 tgatccaca -94 6.87 bhal ttcttcaca -209 6.89 hinf tgatccaca -323 7.01 hinf ttatccaat -100 6.26 vchol ttattcaca -94 8.38 • Psd catalyzes last step of PE synthesis • psd knockouts nonmotile Cronan Jr, J.E. et al., Escherichia coli and Salmonella typhimurium: cellular and molecular biology, F. Neidhardt, et al., Editors.1996, American Society for Microbiology: Washington DC. p. 620. phosphatidylglycerophosphate synthase (pgsA) uvrY-uvrC-pgsA Site Sequence Position Score ecolik12 ttgtccaca -130 7.04 stlt2 ttacccaca -337 6.91 ypes ttctccaga -7 6.73 • pgsA encodes phosphatidylglycerophosphate synthase • catalyzes the committed step of PG and CL biosynthesis in E. coli Cronan Jr, J.E. et al., Escherichia coli and Salmonella typhimurium: cellular and molecular biology, F. Neidhardt, et al., Editors.1996, American Society for Microbiology: Washington DC. p. 620. Conclusion Conclusion • DnaA regulation may couple lipid cellular processes to DNA replication – this may be accomplished by the transcription factor activity of DnaA upstream of phospholipid biosynthesis genes fadL, acpP, fabF, accD, plsC, psd, and pgsA – changes in expression of the phospholipid biosynthesis proteins alter the fatty acid composition of the cell membrane – interactions between DnaA protein and the membrane that modulate the activity of the mutant DnaA • The biological significance of the motifs presented here will be verified experimentally – microarray analysis of temperature sensitive dnaA mutants in a variety of bacteria – chromatin immunoprecipitation studies to verify true positive candidate binding sites Lodish et al., Molecular Cell Biology, 2000, Freeman: NY. p 459. Acknowledgements • • • • Simon Kasif (Bioinformatics, Boston University) Alan Grossman (Microbiology, Massachusetts Institute of Technology) Tohru Mizushima (Pharmaceutical Sciences, Kyushu University ) NSF (KDI) & GEM fellowship Comparative genomic analysis of lipid biosynthesis and metabolism components of the DnaA regulon. Genome Biology. In review.