* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Chapter 4
Oligonucleotide synthesis wikipedia , lookup
RNA interference wikipedia , lookup
RNA silencing wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Ribosomally synthesized and post-translationally modified peptides wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Point mutation wikipedia , lookup
Polyadenylation wikipedia , lookup
Protein structure prediction wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Peptide synthesis wikipedia , lookup
Metalloprotein wikipedia , lookup
Proteolysis wikipedia , lookup
Biochemistry wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Amino acid synthesis wikipedia , lookup
Gene expression wikipedia , lookup
Messenger RNA wikipedia , lookup
Genetic code wikipedia , lookup
Epitranscriptome wikipedia , lookup
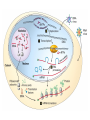
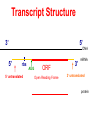
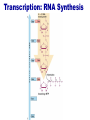
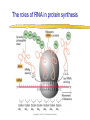
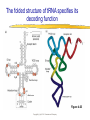
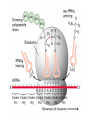
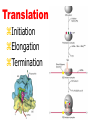
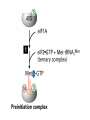
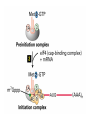
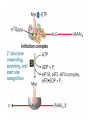
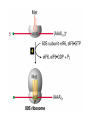
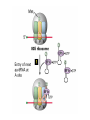
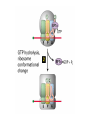
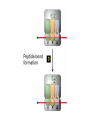
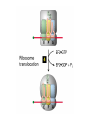
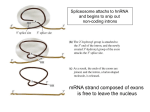
Lecture 3 Gene Structure, Transcription, & Translation Reading: Chapter 4: 108-115; 118-131 Molecular Biology syllabus web site Typical Gene Structure Promoter Coding Region +1 transcription Prokaryotes COORDINATED GENE EXPRESSION: clustered genes (operon) controlled by one promoter and transcribed as polycistronic mRNA and encode multiple gene products Eukaryotes Interrupted genes (exons/introns) Monocistronic mRNAs Post-transcriptional modifications (nuclear encoded genes): 5’ CAP polyA tail splicing Post-transcription addition of 5’ CAP to nuclear encoded eukaryotic mRNA Transcript Structure 3’ 5’ DNA 5’ rbs 5’ untranslated AUG ORF Open Reading Frame 3’ mRNA 3’ untranslated protein Transcription: RNA Synthesis 1. 2. 3. 4. 5. Requirements Enzyme: RNA Polymerase DNA Template (3’ to 5’ strand) No primer required Nucleoside triphosphates: ATP, GTP, CTP, UTP Synthesis is 5’ to 3’ Transcription: RNA Synthesis Translation: Protein Synthesis Codons specify amino acids; positioning on ribosome sets READING FRAME The roles of RNA in protein synthesis Copyright (c) by W. H. Freeman and Company The three roles of RNA in protein synthesis Three types of RNA molecules perform different but complementary roles in protein synthesis (translation) Messenger RNA (mRNA) carries information copied from DNA in the form of a series of three base “words” termed codons Transfer RNA (tRNA) deciphers the code and delivers the specified amino acid Ribosomal RNA (rRNA) associates with a set of proteins to form ribosomes, structures that function as proteinsynthesizing machines Copyright (c) by W. H. Freeman and Company The folded structure of tRNA specifies its decoding function Figure 4-26 Copyright (c) by W. H. Freeman and Company Aminoacyl-tRNA synthetases activate amino acids by linking them to tRNAs Each tRNA molecule is recognized by a specific aminoacyltRNA synthetase Fidelity of protein synthesis determined by: Correct aminoacylation of tRNA Codon-anticodon pairing Aminoacyl tRNA synthetases -at least one for every amino acid -for different codons have different synthetases -error correction lies in specificity of synthetase and tRNA. No mechanism exists for error correction once tRNA is mischarged and separated from synthetase Double sieve mechanism for error correction Synthetases have 2 sites: active site, hydrolytic site. Amino acids larger than the correct amino acid are never activated because they are too large to fit into the active site. Smaller amino acids (than the correct one) fit into the hydrolytic site (which excludes the correct amino acid) and are hydrolyzed. Nonstandard base pairing often occurs between codons and anticodons Ribosomes: the macromolecular site for protein synthesis Translation Initiation Elongation Termination Initiation mRNA binds to ribosome Selection of initiation codon Binding of charged initiator tRNA (first amino acid) Initiation Formation of 30S preinitiation complex 30 S subunit (contains 16S rRNA), mRNA, charged tRNA f-met, initiation factors, GTP + 50S subunit (GTP hydrolysis) Resulting in formation of the 70S initiation complex fmet-tRNA is fixed into the “P site” reading frame is now determined. Initiation Initiation of eukaryotic protein synthesis generally occurs at the 5’ end of mRNA but may occasionally occur at internal sites Initiation of prokaryotic protein synthesis generally occurs at the Shine Delgarno site The untranslated leader or 5’ end of prokaryotic mRNAs contain a ribosome binding site (rbs) or Shine Delgarno site located upstream of the AUG and complementary to the 3’ end of the 16S rRNA. mRNA: 5’ ….AGGAGGU……………..AUG 3’end of 16S rRNA 3’ ...UCCUCCA…………………….. IIIIIII Elongation Peptide bond formation Movement of mRNA/ ribosome (translocation) so each codon may be “read” Elongation Requirements: Elongation factors and GTP hydrolysis Occupation of “A” site by next tRNA Peptide bond formed by peptidyl transferase enzyme Uncharged tRNA-fmet in P site and dipeptidyl tRNA in A site Translocation: deacylated tRNA fmet leaves P site peptidyl tRNA moves from A to P site mRNA moves 3 bases to position next codon at A site Elongation Termination when termination codons are reached (UGA, UAA, UAG) Completed protein is dissociated from machinery Ribosome released Termination when termination codons are reached (UGA, UAA, UAG) Peptidyl tRNA moves from A to P site Release factors (RF) recognize specific stop codons RF forms activated complex with GTP Activated complex binds to termination codon and alters specificity of peptidyl transferase In presence of RF, peptidyl transferase catalyzes reaction of bound peptidyl moiety with water instead of with free aminoacyl tRNA Release of polypeptide Dissociation of 70S ribosome into 50S and 30S subunits. Summary of Protein Synthesis 1. Binding of mRNA to ribosome 2. Charged, amino-acylated initiator tRNA binds to P site of ribosome and is based paired through tRNA anticodon to codon on mRNA 3. A second amino-acylated tRNA fills A site and anticodon Hbonds with second codon on mRNA 4. Amino acids in P and A site are joined by a peptide bond. tRNA in P site is released. tRNA (with 2 amino acids joined) in A site moves to P site A new amino-acylated tRNA moves into A site by anticodon-codon pairing 5. Step (4) is repeated until codon in A site is a stop codon; peptide is released. Post-translational Modifications (Bacteria) removal of formyl groups (fmet) removal of first few amino acids (aminopeptidase) glycosylation (affects targeting, activity) phosphorylation (by kinases) S-S bond formation Polypeptide cleavage -removal of transit peptide upon organelle import -removal of signal sequence (ER secretion) -activation of enzymes