* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Dr. Peter John M.Phil, PhD Assistant Professor National University of
RNA interference wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Designer baby wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Gene expression profiling wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
History of RNA biology wikipedia , lookup
Messenger RNA wikipedia , lookup
Point mutation wikipedia , lookup
Non-coding RNA wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
Gene nomenclature wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Protein moonlighting wikipedia , lookup
Epitranscriptome wikipedia , lookup
RNA-binding protein wikipedia , lookup
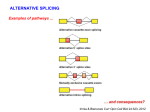
Dr. Peter John M.Phil, PhD Atta-ur-Rahman School of Applied Biosciences (ASAB) National University of Sciences & Technology (NUST) RNA Splicing & Processing Alternative splicing Alternative Splicing: When a single gene give rise to more than one type of mRNA Use of different start points/ generation of alternative 3’ ends alter the pattern of splicing In some cases the multiple products are made in the same cell but in other the splicing is normal Alternative splicing An alternative splicing pathway uses another set of snRNPs that comprise the U12 spliceosome. The target introns are defined by longer consensus sequences at the splice junctions, but usually include the same GU-AG junctions. Some introns have the splice junctions AU-AC, including some that are U1-dependent and some that are U12-dependent. Alternative Splicing in E1A Adeno virus E1A region by using alternative 5’ sites generates different size products T antigen is larger in size t antigen is smaller in size Alternative Splicing in E1A t antigen producing cell From these cell extract protein ASF (Alternative Splicing factor) This protein (ASF) (alternative splicing factor) have same role as splicing factor SF2, required for early steps in spliceosome assembly and for the first cleavage-ligation reaction Alternative Splicing in Mouse Exon 2 & 3 of the Mouse Tropnin T gene are mutantly exclusive Exon-2 used in smooth muscle & Exon -3 used in other tissue Smooth muscle contain protein that binds to repeated elements located on either side of the exon3 & prevent use of the 3’ & 5’ sites that are needed to include it Drosophila Sex Determination Male include exon-3 of sxl gene which have termination ultimately no SXL protein Female exclude exon-3 of sxl gene SXL protein in female SXL protein change the splicing of Tra gene Tra produce Tra2 gene Tra + Tra 2 produce dsx (double sex) gene DSX protein suppress male genes & promote female development Drosophila Sex Determination Sex determination in D. melanogaster involves a pathway in which different splicing events occur in females. Blocks at any stage of the pathway result in male development. Splicing and mRNA export Spliceosome also may provide the initial point of contact for the export apparatus. A protein complex binds to the RNA via the splicing apparatus The complex consists of >9 proteins and is called the EJC (exon junction complex) Exon Junction Complex EJC EJC is involved in several functions Some of the proteins of the EJC are directly involved in association with splicing Factors Others recruit additional proteins for particular functions mRNA Transport EJC includes a group of proteins called the REF family REF proteins in turn interact with a transport protein (called TAP and Mex) TAP and Mex has direct responsibility for interaction with the nuclear pore Thanks