* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Control of Gene Expression
Gene therapy of the human retina wikipedia , lookup
Gene nomenclature wikipedia , lookup
Minimal genome wikipedia , lookup
Genome evolution wikipedia , lookup
Protein moonlighting wikipedia , lookup
X-inactivation wikipedia , lookup
Ridge (biology) wikipedia , lookup
Gene expression programming wikipedia , lookup
Epigenetics wikipedia , lookup
History of genetic engineering wikipedia , lookup
Cancer epigenetics wikipedia , lookup
Transcription factor wikipedia , lookup
Polyadenylation wikipedia , lookup
Genome (book) wikipedia , lookup
History of RNA biology wikipedia , lookup
Epigenetics in stem-cell differentiation wikipedia , lookup
Non-coding DNA wikipedia , lookup
Point mutation wikipedia , lookup
Genomic imprinting wikipedia , lookup
Microevolution wikipedia , lookup
Epigenomics wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Short interspersed nuclear elements (SINEs) wikipedia , lookup
Designer baby wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Epigenetics of neurodegenerative diseases wikipedia , lookup
RNA silencing wikipedia , lookup
Messenger RNA wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Epigenetics in learning and memory wikipedia , lookup
RNA interference wikipedia , lookup
Gene expression profiling wikipedia , lookup
Long non-coding RNA wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Polycomb Group Proteins and Cancer wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Non-coding RNA wikipedia , lookup
Mir-92 microRNA precursor family wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
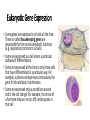
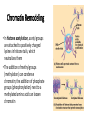
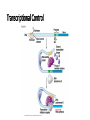
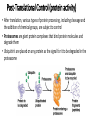
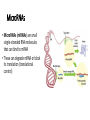
Control of Gene Expression Eukaryotes Eukaryotic Gene Expression • Some genes are expressed in all cells all the time. These so-called housekeeping genes are responsible for the routine metabolic functions (e.g. respiration) common to all cells. • Some are expressed as a cell enters a particular pathway of differentiation. • Some are expressed all the time in only those cells that have differentiated in a particular way. For example, a plasma cell expresses continuously the gene for the antibody it synthesizes. • Some are expressed only as conditions around and in the cell change. For example, the arrival of a hormone may turn on (or off) certain genes in that cell One Gene = One Protein Hypothesis • By Beadle and Tatum (1940’s) • The one gene-one enzyme hypothesis is the idea that genes act through the production of enzymes, with each gene responsible for producing a single enzyme that in turn affects a single step in a metabolic pathway. • http://www.dnalc.org/view/163 60-Animation-16-One-genemakes-one-protein-.html The Eukaryotic Gene • Associated with most eukaryotic genes are control elements, segments of noncoding DNA that help regulate transcription by binding certain proteins • Control elements and the proteins they bind are critical to the precise regulation of gene expression in different cell types Many Levels of Control in Eukaryotes • Chromatin Remodeling The region of the chromosome must be opened up in order for enzymes and transcription factors to access the gene • Transcription Control The most common type of genetic regulation • Turning on and off of mRNA formation • Post-Transcriptional Control Regulation of the processing of a pre-mRNA into a mature mRNA • Translational Control Regulation of the rate of Initiation • Post-Tranlational Control (protein activity control) Regulation of the modification of an immature or inactive protein to form an active protein Chromatin Remodeling • Genes within highly packed heterochromatin are usually not expressed • Chemical modifications to histones and DNA of chromatin influence both chromatin structure and gene expression • Requires ATP Chromatin Remodeling • In histone acetylation, acetyl groups are attached to positively charged lysines in histone tails, which neutralizes them • The addition of methyl groups (methylation) can condense chromatin; the addition of phosphate groups (phosphorylation) next to a methylated amino acid can loosen chromatin Chromatin Remodeling Transcription Control • To initiate transcription, eukaryotic RNA polymerase requires the assistance of proteins called transcription factors • In eukaryotes, high levels of transcription of particular genes depend on control elements interacting with specific transcription factors • Proximal control elements are located close to the promoter • Distal control elements, groups of which are called enhancers, may be far away from a gene or even located in an intron • An activator is a protein that binds to an enhancer and stimulates transcription of a gene Transcriptional Control Transcriptional Control • Methylation of bases also turns off transcription • DNA methylation can cause long-term inactivation of genes in cellular differentiation • In genomic imprinting, methylation regulates expression of either the maternal or paternal alleles of certain genes at the start of development • Although the chromatin modifications just discussed do not alter DNA sequence, they may be passed to future generations of cells • The inheritance of traits transmitted by mechanisms not directly involving the nucleotide sequence is called epigenetic inheritance Post-Transcriptional Control • Alternative RNA splicing: different mRNA molecules are produced from the same primary transcript, depending on which exons are spliced out by the splicosome • mRNA degradation: without a 5’ cap and poly-A tail an mRNA will be destroyed Post-Translational Control (protein activity) • After translation, various types of protein processing, including cleavage and the addition of chemical groups, are subject to control • Proteasomes are giant protein complexes that bind protein molecules and degrade them • Ubiquitin's are placed on any protein as the signal for it to be degraded in the proteasome Noncoding RNAs • Only a small fraction of DNA codes for proteins, rRNA, and tRNA • A significant amount of the genome may be transcribed into noncoding RNAs • Noncoding RNAs regulate gene expression at two points: mRNA translation and chromatin configuration MicroRNAs • MicroRNAs (miRNAs) are small single-stranded RNA molecules that can bind to mRNA • These can degrade mRNA or block its translation (translational control) siRNAs • The phenomenon of inhibition of gene expression by RNA molecules is called RNA interference (RNAi) • RNAi is caused by small interfering RNAs (siRNAs) • siRNAs play a role in heterochromatin formation and can block large regions of the chromosome • Small RNAs may also block transcription of specific genes miRNA vs. siRNA • both are formed from dsRNA and both eventually get cleaved into pieces by Dicer and then incorporated into RISC which in effect cleaves target mRNA. • siRNAs and miRNAs are similar but form from different RNA precursors • Sirna Therapeutics, Inc. was a San Francisco, California based biotechnology company that explores the use of RNA interference in human disease therapy. Sirna's development pipeline includes several small interfering RNA (siRNA) drugs, thought to stably silence the expression of specific disease-related genes.