* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Development of a molecular genetic diagnostic service for X
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Genetic testing wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Tay–Sachs disease wikipedia , lookup
Metagenomics wikipedia , lookup
Genetic engineering wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Public health genomics wikipedia , lookup
Genome evolution wikipedia , lookup
Gene desert wikipedia , lookup
Gene expression programming wikipedia , lookup
Population genetics wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Genome (book) wikipedia , lookup
Gene expression profiling wikipedia , lookup
Gene therapy wikipedia , lookup
Gene nomenclature wikipedia , lookup
Oncogenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Neuronal ceroid lipofuscinosis wikipedia , lookup
Genome editing wikipedia , lookup
Helitron (biology) wikipedia , lookup
Saethre–Chotzen syndrome wikipedia , lookup
Frameshift mutation wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Medical genetics wikipedia , lookup
Designer baby wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
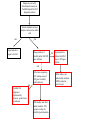
Development of a molecular genetic diagnostic service for X-linked ichthyosis, with emphasis on carrier detection Eleanor Reavey West of Scotland Regional Genetics Laboratory Yorkhill Hospital Glasgow Introduction Associated with STS deficiency in fibroblasts and ↑ plasma cholesterol sulphate X-linked recessive inheritance 1 in 2000-6000 males STS gene - Xp22.3 10 exons STS enzyme Responsible for hydrolysis of cholesterol sulfate (CS) to cholesterol in epidermis XLI – accumulation of CS in epidermis leads to barrier instability and inhibits desmosomal degradation Phenotype Scaly skin on scalp, trunk and limbs Corneal opacities Placental sulphatase deficiency Placenta – STS-rich tissue STS is involved in steroid conversion pathway: cholesterol estriol Deficiency associated with: longer gestation and poor cervical dilatation Results in slowing of delivery + indicates need for C-section or instrumental delivery ↑ perinatal morbidity + mortality Associated Conditions Approx. 90% of XLI individuals – complete deletion of the STS gene More extensive deletions - contiguous gene deletion syndromes Kallmann syndrome Short stature X-linked chondrodysplasia punctata X-linked ocular albinism ADHD Biochemical Analysis STS activity is measured on white cells or cultured fibroblasts Radiolabelled assay with 3H Dehydroepiandrosterone sulphate as a substrate Affected males are tested for presence or absence of STS gene by PCR No info on any intragenic deletions or point mutations Mutations Several point mutations in STS gene identified No evidence of genotype-phenotype correlation, regardless of the location or type of the STS mutation production of a catalytically inactive STS enzyme both the N-terminal region and the C-terminal region of the STS protein are important for enzyme activity Initial referral Patient NH clinically affected with XLI No enzyme activity detected But, normal result for gene deletion analysis Request from Dundee for full seq screen of STS coding exons (1-10), including intron/exon boundaries Primers designed for sequencing Y chr Pseudogene Transcriptionally inactive at the promoter Several exons deleted Significant sequence homology between X-STS and Y-STS genes Results from Temperature Gradient PCR 55°C - 65°C Example gel for exons 1-8 Exon 1 2 3 4 5 6 7 8 55C 58C 60C 62C 65C NH c.583delG p.Val195SerfsX19 Further Testing Screening of NH’s mother confirmed her as a carrier. Second referral – Edinburgh Patient JM clinically affected, no STS activity and normal result on gene deletion analysis JM c.387_391dupAGCAC p.Leu131GlnfsX3 Extended Testing A further 10 samples were received from Dr Graham Dosage analysis carried out to confirm presence of STS gene Full sequencing screen carried out on all 10 exons Four additional mutations detected = high pickup rate STS dosage analysis DMD 53 STS 5’ DMD 17 STS 3’ DMD 51 MD c.1046_1048delAGG p.Glu349del TT c.1649G>A p.Trp550X AE c.1360C>T p.Arg454Cys JA c.494C>T p.Thr165Ile MLPA kit P160 Probes for each of 10 exons Other probes include KAL1 and NLGN4X In female heterozygotes, 35-50% reduced relative peak area of amplified product expected Deletion of one exon – needs to be confirmed by sequencing to rule out mutation/ polymorphism close to probe ligation site Carrier testing in females Current testing strategy for XLI in Glasgow Enzyme activity measured and gene deletion PCR carried out in Biochemical Genetics Dosage assay available in Molecular Genetics Lab to identify female carriers MLPA better suited for carrier testing – detects single (or multiple) exon deletions/ duplications as well as deletions of entire gene Sample is received by Biochemical Genetics at Yorkhill Hospital for XLI diagnostic analysis Steroid sulphatase enzyme analysis carried out on white cells -ve +ve Report patient as negative for XLI Dosage analysis to identify partial/ full STS gene deletions +ve Report patient is affected with XLI due to a STS gene deletion -ve Full screen sequencing of 10 coding exons of STS gene to identify point mutations -ve Confirm XLI diagnosed biochemically however, genetic basis is unknown Offer mother, and other family members, STS sequence testing for identified point mutation Offer mother, and other family members, MLPA testing for carrier status Summary Service offered for males affected with XLI – dosage analysis + full screen sequencing for point mutations Carrier testing for mothers Important for genetic counselling for future pregnancies and for predicting risk of difficult labour Acknowledgements Molecular Genetics Lab, Yorkhill, Glasgow Su Stenhouse, Sandy Cooke Biochemical Genetics Lab, Yorkhill, Glasgow Gordon Graham