* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Mapping QTL and genes in tilapias
Cancer epigenetics wikipedia , lookup
Copy-number variation wikipedia , lookup
Epigenetics of diabetes Type 2 wikipedia , lookup
Metagenomics wikipedia , lookup
Biology and consumer behaviour wikipedia , lookup
Extrachromosomal DNA wikipedia , lookup
Mitochondrial DNA wikipedia , lookup
Cell-free fetal DNA wikipedia , lookup
Ridge (biology) wikipedia , lookup
Gene desert wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Point mutation wikipedia , lookup
Epigenetics of human development wikipedia , lookup
Genealogical DNA test wikipedia , lookup
Genomic imprinting wikipedia , lookup
Transposable element wikipedia , lookup
Genetic engineering wikipedia , lookup
Oncogenomics wikipedia , lookup
Therapeutic gene modulation wikipedia , lookup
Gene expression profiling wikipedia , lookup
Whole genome sequencing wikipedia , lookup
No-SCAR (Scarless Cas9 Assisted Recombineering) Genome Editing wikipedia , lookup
Nutriepigenomics wikipedia , lookup
Pathogenomics wikipedia , lookup
Genome (book) wikipedia , lookup
Designer baby wikipedia , lookup
Public health genomics wikipedia , lookup
Site-specific recombinase technology wikipedia , lookup
Non-coding DNA wikipedia , lookup
Human genome wikipedia , lookup
Helitron (biology) wikipedia , lookup
Microevolution wikipedia , lookup
Genomic library wikipedia , lookup
History of genetic engineering wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Human Genome Project wikipedia , lookup
Microsatellite wikipedia , lookup
A30-Cw5-B18-DR3-DQ2 (HLA Haplotype) wikipedia , lookup
Minimal genome wikipedia , lookup
Genome editing wikipedia , lookup
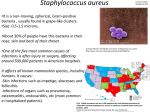
Mapping genes and QTL in tilapias Gideon Hulata1, Avner Cnaani2, Bo-Young Lee2, Woo-Jai Lee3, Aimee E. Howe2, J. Todd Streelman2, and Thomas D. Kocher2 1 Department of Aquaculture, Institute of Animal Science Agricultural Research Organization, Volcani Center, PO Box 6 Bet Dagan 50250 Israel 2 Hubbard Center for Genome Studies, University of New Hampshire Durham, NH 03824 USA 3 GenoMar ASA, Oslo Research Park, Gaustadalleen 21 Oslo, 0349 Norway The ability of tilapiine fishes to create viable interspecific hybrids makes them an ideal organism for genetic studies, using backcrosses or F2 intercrosses as a segregating population. Several linkage maps of DNA markers were constructed for tilapias in recent years. The recent development of hundreds of microsatellite DNA markers enable coverage of the tilapia genome at 2.4 cM intervals on average, thus providing the infrastructure for systematic genome scans for detection of QTL. Markers associated with genes with deleterious alleles and distorted sex ratios were found in an inbred gynogenetic line of Oreochromis aureus. Variation in a microsatellite was found to be associated with expression of the prolactin gene and growth in salinity-challenged fish from the F2 of an O. mossambicus x O. niloticus cross. A locus causing lack of melanin in a red tilapia was localized to LG3 through analysis of color segregation in two F2 hybrid populations (O. aureus x Red O. niloticus and Red O. mossambicus x O. niloticus). Trp1, a gene essential to melanin synthesis, was mapped to the same region, but is not the causative gene for this mutation. Chromosomal regions strongly associated with sex determination were localized to LG1 (Y haplotype) in O. niloticus, and both LG1 (Y haplotype) and LG3 (W haplotype) in O. aureus. Analysis of epistatic interactions among the loci suggests the action of a dominant male repressor (the W haplotype on LG 3) and a dominant male determiner (the Y haplotype on LG1). A locus associated with sex determination in Tilapia zillii was also localized to LG1, yet to a different region than the one in O. aureus and O. niloticus. A chromosomal region on LG23 with putative QTL for cold tolerance and body weight was detected by a preliminary scan of an F2 hybrid population involving two species of tilapia, O. aureus and O. mossambicus, differing in their tolerance to low temperature, their innate immune response to stress and several biochemical blood components, and their growth rate. The region was re-sampled in a second experiment, confirming the results of the first experiment. These results were further supported by the results of another genome scan performed on a four-way cross between a (O. niloticus x S. galilaeus) male and a (O. mossambicus x O. aureus) female. Genome scan using 42 DNA markers, covering ~80% of the tilapia genome, performed on another family of the O. mossambicus x O. aureus F2 hybrid population revealed markers association with stress response, body weight and sex determination in four linkage groups: LG 1, 3, 4 and 23, confirming the location of QTL reported by several other studies. Two out of the 42 markers utilized in this study were polymorphic genes, MHC class I (MHC-1) and Transferrin (TF), which are related to the immune system, and previously mapped in the tilapia genome. In addition, two of the microsatellite markers UNH881 and UNH208 were previously found to be part of coding genes of the IgM light chain and Attractin, respectively.