* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download concentration gradient
Survey
Document related concepts
P-type ATPase wikipedia , lookup
Magnesium transporter wikipedia , lookup
Signal transduction wikipedia , lookup
Implicit solvation wikipedia , lookup
Mechanosensitive channels wikipedia , lookup
SNARE (protein) wikipedia , lookup
Membrane potential wikipedia , lookup
Theories of general anaesthetic action wikipedia , lookup
List of types of proteins wikipedia , lookup
Lipid bilayer wikipedia , lookup
Cell membrane wikipedia , lookup
Transcript
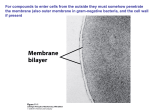
Molecular, Cellular and Developmental Neuroscience January 10, 2008 9-10:50 am Membranes and Membrane Proteins Lecturer: Professor Eileen M. Lafer Contact Info.: 415B, 567-3764, [email protected] Recommended Reading: Stryer Edition 5: Chapters 12 and 13, pp. 319-369 or Stryer Edition 6: Chapters 12 and 13, pp. 326-372 BIOLOGICAL MEMBRANES 1. The boundaries of cells are formed by biological membranes. 2. The boundaries of organelles are also formed by biological membranes. 3. Membranes define inside and outside of a cell or organelle. 4. Membranes confer cells and organelles with selective permeability. CHEMICAL SYNAPSE MACROMOLECULAR CONSTITUENTS OF MEMBRANES 1. LIPIDS: cholesterol sphingolipids: sphingomyelin (SP); gangliosides glyceryl phospholipids: phosphatidylcholine (PC), phosphatidlyethanolamine (PE), phosphatidylglycerol (PG), phosphatidlyserine (PS), phosphatidlyinositol (PI), cardiolipin (CL). 2. PROTEIN: integral and peripheral. 3. CARBOHYDRATE: in the form of glycoprotein and glycolipid, never free. BIOLOGICAL LIPIDS ARE AMPHIPATHIC Hydrophobic Hydrophilic Tail Head HOW DO AMPHIPATHIC MOLECULES ARRANGE THEMSELVES IN AQUEOUS SOLUTIONS? 1. Micelles: limited structures microscopic dimensions: <20 nm in diameter MOVIE: MICELLE FORMATION MemSnD1.avi 2. Lipid Bilayers: bimolecular sheet macroscopic dimensions: 1 mm = 106 nm MOVIE: BILAYER FORMATION MemSnD2.avi Both of these arrangements allow the hyrdrophobic regions to be shielded from the aqueous environment, while the hydrophilic regions are in contact with the aqueous environment. Which arrangement is favored by biological lipids? BILAYERS The two fatty acyl chains of a phospholipid or glycolipid are too bulky too fit in the interior of a micelle. LIPID BILAYERS FORM BY SELF-ASSEMBLY 1. The structure of a bimolecular sheet is inherent in the structure of the constituent lipid molecules. 2. The growth of lipid bilayers from phospholipids is a rapid and spontaneous process in aqueous solution. LIPID BILAYERS ARE COOPERATIVE STRUCTURES 1. They are held together by many reinforcing non-covalent interactions, which makes them extensive. 2. They close on themselves so there are no edges with hydrocarbon chains exposed to water, which favors compartmentalization. 3. They are self-sealing because a hole is energetically unfavorable. CHEMICAL FORCES THAT STABILIZE LIPID BILAYERS 1. Hydrophobic interactions are the primary force. These occur between the extensive hydrophobic lipid tails that are stacked in the sheet. 2. van der Waals attractive forces between the hydrocarbon tails favor their close packing. 3. Electrostatic interactions lead to hydrogen bond formation between the polar head groups and water molecules in the solution. THEREFORE THE SAME CHEMICAL FORCES THAT STABILIZE PROTEIN STRUCTURES STABILIZE LIPID BILAYERS MEMBRANES THAT PERFORM DIFFERENT FUNCTIONS CONTAIN DIFFERENT SETS OF PROTEINS A. Plasma membrane of an erythrocyte. B. Photoreceptor membrane of a retinal rod cell. C. Sarcoplasmic reticulum membrane of a muscle cell. PROTEINS ASSOCIATE WITH THE LIPID BILAYER IN MANY WAYS 1. A,B,C: Integral membrane proteins -Interact extensively with the bilayer. -Require a detergent or organic solvent to solubilize. 2. D,E: Peripheral membrane proteins -Loosely associate with the membrane, either by interacting with integral membrane proteins or with the polar head groups of the lipids. -Can be solubilized by mild conditions such as high ionic strength. PROTEINS CAN SPAN THE MEMBRANE WITH ALPHA HELICES Structure of bacteriorhodopsin. MOST COMMON STRUCTURAL MOTIF a-helices are composed of hydrophobic amino acids. Cytoplasmic loops and extracellular loops are composed of hydrophilic amino acids. A CHANNEL PROTEIN CAN BE FORMED BY BETA SHEETS Structure of bacterial porin. Hydrophobic amino acids are found on the outside of the pore. Hydrophilic amino acids line the aqueous central pore. INTEGRAL MEMBRANE PROTEINS DO NOT HAVE TO SPAN THE ENTIRE LIPID BILAYER Prostaglandin H2 synthase-1 (one monomer of dimeric enzyme is shown) PROTEIN DIMERIZATION LEADS TO THE FORMATION OF A HYDROPHOBIC CHANNEL IN THE MEMBRANE PERIPHERAL MEMBRANE PROTEINS CAN ASSOCIATE WITH MEMBRANES THROUGH COVALENTLY ATTACHED HYDROPHOBIC GROUPS FLUID MOSAIC MODEL ALLOWS LATERAL MOVEMENT BUT NOT ROTATION THROUGH THE MEMBRANE LIPID MOVEMENT IN MEMBRANES LIPIDS UNDERGO A PHASE TRANSITION WHICH FACILITATES THE LATERAL DIFFUSION OF PROTEINS MOVIE: LIPID DYNAMICS MemSnD3.avi MOVIE: FLUID MOSAIC MODEL MemSnD4.avi MEMBRANE FLUIDITY IS CONTROLLED BY FATTY ACID COMPOSITION AND CHOLESTEROL CONTENT DIFFUSION ACROSS A MEMBRANE The rate of diffusion of any molecule across a membrane is proportional to BOTH the molecule's diffusion coefficient and its concentration gradient. 1. The diffusion coefficient (D) is mainly a function of the lipid solubility of the molecule. Hydrophilic molecules (water soluble, e.g. sugars, charged ions) diffuse more slowly. Hydrophobic molecules (e.g. steroids, fatty acids) diffuse more rapidly. 2. The concentration gradient (Cside1/Cside2) is the difference in concentration across the membrane. Diffusion always occurs from a region of higher concentration to a region of lower concentration. For any given molecule, the greater the concentration difference the greater the rate of diffusion. THE DIRECTION OF DIFFUSION FOLLOWS THE CONCENTRATION GRADIENT OUT IN 10 mM 1 mM 5 mM 1 mM no diffusion 5 mM 10 mM DIFFUSION RATES The overall rate of diffusion is determined by multiplying the diffusion coefficient and the magnitude of the concentration gradient: Rate ~ D x (Cside1/Cside2) SMALL LIPOPHILIC MOLECULES DIFFUSE ACROSS MEMBRANES BY SIMPLE DIFFUSION 1. The small molecule sheds its solvation shell of water. 2. Then it dissolves in the hydrocarbon core of the membrane. 3. Then it diffuses through the core to the other side of the membrane along its concentration gradient. 4. Then it is resolvated by water. MOVIE: LARGE AND POLAR MOLECULES DO NOT READILY DIFFUSE ACROSS MEMBRANES BY SIMPLE DIFFUSION MemIT1.avi LARGE AND POLAR MOLECULES ARE TRANSPORTED ACROSS MEMBRANES BY PROTEINACEOUS MEMBRANE TRANSLOCATION SYSTEMS 1. Passive Transport (also called facilitated diffusion): The transport goes in the same direction as the concentration gradient. This does not require an input of energy. 2. Active Transport: The transport goes in the opposite direction as the concentration gradient. This requires an input of energy. SODIUM-POTASSIUM PUMP Actively exchanges sodium and potassium against their concentration gradients utilizing the energy of ATP hydrolysis. MOVIE: Establishes the concentration gradients of sodium and potassium essential for synaptic transmission. Movie 12-10.avi ACETYLCHOLINE RECEPTOR Passively transports sodium and potassium ions along their concentration gradients in response to neuronal signals. Example of a ligand-gated ion channel. MOVIE: Movement through an ion channel. MemIT4.avi CLASSIFICATION OF MEMBRANE TRANSLOCATION SYSTEMS: TYPE CLASS Channel (translocates ~107 ions per second) Passive Transport 1. Voltage Gated 2. Ligand Gated 3. cAMP Regulated 4. Other EXAMPLE Na+ channel AChR Cl- channel Pressure Sensitive Transporter (translocates ~102-103 molecules per second) Passive Transport Glucose transporter Active Transport 1. 1o-ATPase Na+/K+ Pump 2. 1o-redox coupled Respiratory Chain Linked 3. ATP-binding Multidrug resistance cassette protein transporter 4. 2o Na+-dependent glucose transport ACTIVE TRANSPORT MECHANISMS 1o 2o Utilizes the energy of ATP Utilizes the downhill flow hydrolysis to transport a of one gradient to power molecule against its the formation of another concentration gradient. gradient. MEMBRANE TRANSLOCATION SYSTEMS DIFFER IN THE NUMBER AND DIRECTIONALITY OF THE SOLUTES TRANSPORTED This classification is independent of whether the transport is active or passive. EXAMPLE: Uniporter Primary Active TransportTransports Ca++ against its concentration gradient utilizing the energy of ATP hydrolysis Sarcoplasmic Reticulum Ca++ ATPase MECHANISM of SR Ca++ ATPase EXAMPLE: Symporter Secondary Active Transport-Na+-Glucose Symporter: Transports glucose against its concentration gradient utilizing the downhill flow of Na+ along its concentration gradient previously set up by the Na+/K+ pump. Na+/K+ Pump Na+-Glucose Symporter EXAMPLE: Uniporter Passive Transport Voltage Gated Ion Channel K+ Channel: selectively transports K+ PATH THROUGH THE K+ CHANNEL MECHANISM OF ION SELECTIVITY OF THE K+ CHANNEL K+ ions interact with the carbonyl groups of the TVGYG sequence in the selectivity filter. Note, Na+ ions are too small to make sufficient productive interactions with the channel. MECHANISM OF VOLTAGE GATING OF THE K+ CHANNEL "Ball and Chain Mechanism" SUMMARY OF TRANSPORT TYPES