* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Ab initio modelling tutorial (part II)
Survey
Document related concepts
Implicit solvation wikipedia , lookup
X-ray crystallography wikipedia , lookup
Protein domain wikipedia , lookup
Protein mass spectrometry wikipedia , lookup
Protein purification wikipedia , lookup
Western blot wikipedia , lookup
Intrinsically disordered proteins wikipedia , lookup
Homology modeling wikipedia , lookup
Nuclear magnetic resonance spectroscopy of proteins wikipedia , lookup
Protein structure prediction wikipedia , lookup
Transcript
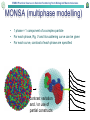
EMBO Practical Course on Solution Scattering from Biological Macromolecules Ab initio tutorial (part II) • Ambimeter – model independent ambiguity score • Damclust – alternative to Damaver • Gasbor (local & online) – real/reciprocal space • Monsa (online) – Nucleoprotein – Dimeric complex • Dammin – limited search volume – anisometry restraint AMBIMETER: tool for Ambiguity Assessment Map of renormalized scattering profiles from exhaustive set of shape topologies is employed to quantitatively evaluate the ambiguity of a small-angle-scattering curve Map of SAXS profiles density Extreme cases 0.0 Flat Disc Rod Sphere lg(I/I0) 0 10 100 1000 10000 -0.4 -0.8 -1.2 -1.6 1 2 3 4 AMBIMETER is now a part of SASFLOW pipeline 5 s*Rg 6 18 Skeletons with identical SAXS curves DamClust: Assessment of multimodality (has damaver & friends inside) Creates the complete graph by iteratively• joining the clusters (singles) Selects the optimal threshold as a compromise between the number of clusters and averaged spread within the cluster Clustering of multiple SAS models – Discrepancies (distances) between multiple models as criteria for grouping – Normalized spatial deviation serves as a distance between heterogeneous models (e.g. bead models) – R.m.s.d. is employed for those with atom-to-atom correspondence (e.g. rigid body models) EMBO Practical Course on Solution Scattering from Biological Macromolecules Dummy Residues Model • Proteins typically consist of folded polypeptide chains composed of amino acid residues • At a resolution of 0.5 nm each amino acid can be represented as one entity (dummy residue) • In GASBOR a protein is represented by an ensemble of K DRs those are – Identical – Have no ordinal number – For simplicity are centered at the Cα positions EMBO Practical Course on Solution Scattering from Biological Macromolecules n: 0 1 2 3 4 5 6 Fib(n): 1 1 2 3 5 8 13 21 34 55 89 144 233 377 610 987 7 ∆ρ=0.03 e/A3 Width=3A ρ=0.334 e/A3 8 9 10 11 12 13 14 15 EMBO Practical Course on Solution Scattering from Biological Macromolecules MONSA (multiphase modelling) • 1 phase = 1 component of a complex particle • For each phase, Rg, V and its scattering curve can be given • For each curve, contrast of each phase are specified contrast variation and / or use of partial constructs EMBO Practical Course on Solution Scattering from Biological Macromolecules Monsa case1: protein-RNA complex 252 AA protein (two domains) 67 nucleotides RNA Three curves in total: Free RNA Complex Free protein EMBO Practical Course on Solution Scattering from Biological Macromolecules Monsa case2: dimeric compex • 172 AA (20kDa) and 538 AA (70 kDa) proteins • Dimerization via small one • Two-fold symmetry axis EMBO Practical Course on Solution Scattering from Biological Macromolecules Dammin features missing in Dammif • Specific (limited) search volume (sphere, ellipsoid, cylinder, parallelepiped, User supplied) • Icosahedral and cubic symmetries • Anisometry restraint EMBO Practical Course on Solution Scattering from Biological Macromolecules Anisometry (direction) • both shapes are symmetric and extended