* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Translation - CS
Protein moonlighting wikipedia , lookup
History of molecular evolution wikipedia , lookup
Eukaryotic transcription wikipedia , lookup
Polyadenylation wikipedia , lookup
RNA interference wikipedia , lookup
Cre-Lox recombination wikipedia , lookup
RNA silencing wikipedia , lookup
RNA polymerase II holoenzyme wikipedia , lookup
Gene regulatory network wikipedia , lookup
Genetic code wikipedia , lookup
Community fingerprinting wikipedia , lookup
Gene expression profiling wikipedia , lookup
Ancestral sequence reconstruction wikipedia , lookup
Nucleic acid analogue wikipedia , lookup
Point mutation wikipedia , lookup
Promoter (genetics) wikipedia , lookup
Genome evolution wikipedia , lookup
Messenger RNA wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Deoxyribozyme wikipedia , lookup
Two-hybrid screening wikipedia , lookup
Non-coding RNA wikipedia , lookup
Non-coding DNA wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Epitranscriptome wikipedia , lookup
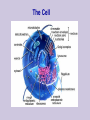
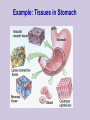
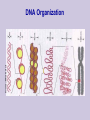
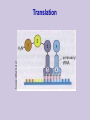
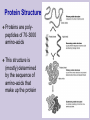
Class 1: Introduction . Source: Alberts et al The Tree of Life The Cell Example: Tissues in Stomach DNA Components Four nucleotide types: Adenine Guanine Cytosine Thymine Hydrogen bonds: A-T C-G Source: Alberts et al The Double Helix Source: Mathews & van Holde DNA Duplication Source: Alberts et al DNA Organization Genome Sizes E.Coli (bacteria) Yeast (simple fungi) Smallest human chromosome Entire human genome 4.6 x 106 bases 15 x 106 bases 50 x 106 bases 3 x 109 bases Genes The DNA strings include: Coding regions (“genes”) E. coli has ~4,000 genes Yeast has ~6,000 genes C. Elegans has ~13,000 genes Humans have ~32,000 genes Control regions These typically are adjacent to the genes They determine when a gene should be expressed “Junk” DNA (unknown function) Transcription sequences can be transcribed to RNA Source: Mathews & van Holde Coding RNA nucleotides: Similar to DNA, slightly different backbone Uracil (U) instead of Thymine (T) RNA Editing Source: Mathews & van Holde RNA Editing RNA roles Messenger RNA (mRNA) Encodes protein sequences Transfer RNA (tRNA) Adaptor between mRNA molecules and aminoacids (protein building blocks) Ribosomal RNA (rRNA) Part of the ribosome, a machine for translating mRNA to proteins ... Transfer RNA Anticodon: matches a codon (triplet of mRNA nucleotides) Attachment site: matches a specific amino-acid Translation Translation is mediated by the ribosome Ribosome is a complex of protein & rRNA molecules The ribosome attaches to the mRNA at a translation initiation site Then ribosome moves along the mRNA sequence and in the process constructs a poly-peptide When the ribosome encounters a stop signal, it releases the mRNA. The construct poly-peptide is released, and folds into a protein. Source: Alberts et al Translation Source: Alberts et al Translation Source: Alberts et al Translation Source: Alberts et al Translation Source: Alberts et al Translation Genetic Code Protein Structure Proteins are polypeptides of 70-3000 amino-acids This structure is (mostly) determined by the sequence of amino-acids that make up the protein Protein Structure Evolution Related organisms have similar DNA Similarity in sequences of proteins Similarity in organization of genes along the chromosomes Evolution plays a major role in biology Many mechanisms are shared across a wide range of organisms During the course of evolution existing components are adapted for new functions Evolution Evolution of new organisms is driven by Diversity Different individuals carry different variants of the same basic blue print Mutations The DNA sequence can be changed due to single base changes, deletion/insertion of DNA segments, etc. Selection bias Course Goals Computational We tools in molecular biology will cover computational tasks that are posed by modern molecular biology We will discuss the biological motivation and setup for these tasks We will understand the the kinds of solutions exist and what principles justify them Four Aspects Biological What is the task? Algorithmic How to perform the task at hand efficiently? Learning How to adapt parameters of the task form examples Statistics How to differentiate true phenomena from artifacts Example: Sequence Comparison Biological Evolution preserves sequences, thus similar genes might have similar function Algorithmic Consider all ways to “align” one sequence against another Learning How do we define “similar” sequences? Use examples to define similarity Statistics When we compare to ~106 sequences, what is a random match and what is true one Topics I Dealing with DNA/Protein sequences: Genome projects and how sequences are found Finding similar sequences Models of sequences: Hidden Markov Models Transcription regulation Protein Families Gene finding Topics II Gene Expression: Genome-wide expression patterns Data organization: clustering Reconstructing transcription regulation Recognizing and classifying cancers Topics III Models of genetic change: Long term: evolutionary changes among species Reconstructing evolutionary trees from current day sequences Short term: genetic variations in a population Finding genes by linkage and association Topics IV Protein World: How proteins fold - secondary & tertiary structure How to predict protein folds from sequences data alone How to analyze proteins changes from raw experimental measurements (MassSpec) 2D gels Class Structure 2 weekly meeting Class: Mondays 16-18 Targil: Tuesday 18-20 Grade: 60% in five question sets Each contains theoretical problems & practical computer questions 40% test 5% bonus for active participation Exercises & Handouts Check regularly http://www.cs.huji.ac.il/~cbio