* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Titel: Functional replacement of Gfi1 deficiency by Gfi1b obviously
Survey
Document related concepts
RNA silencing wikipedia , lookup
X-inactivation wikipedia , lookup
Transcriptional regulation wikipedia , lookup
Non-coding RNA wikipedia , lookup
Gene regulatory network wikipedia , lookup
Epitranscriptome wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
List of types of proteins wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Gene expression profiling wikipedia , lookup
DNA vaccination wikipedia , lookup
Endogenous retrovirus wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Western blot wikipedia , lookup
Monoclonal antibody wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Transcript
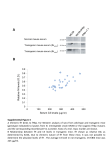
Supplementary material, MS EMBO/2005/82698 third revision Supplementary information Materials and Methods Generation of Gfi:Gfi1b and Gfi1:Gfi1P2A knock-in mice Both knock-in mice were generated by modifying the previously described Gfi1:GFP knockin construct (Yücel et al., 2004). The constructs were transfected in R1 ES cells and selection with G418 and ganciclovir was performed. Mice were held under specific pathogen-free conditions in individually ventilated cages. All animal experiments were carried out according to the German Animal Protection Law and were done under a license granted by the Bezirksregierung Düsseldorf/NRW Germany (Nr.G022/02Z) and the Animal Care and Use Committee of Tel Aviv University (M-04-014). ES cells in which a homologous recombination event occurred were identified by Southern blotting (supp. Fig. 1A and 2A) and were injected into mouse blastocysts to generate Gfi1 +/1b or Gfi1P2A/P2A mutants. The functionality of the newly introduced Gfi1:Gfi1b knock-in allele was confirmed by expression analysis of Gfi1 and Gfi1b in thymocytes (supp. Fig. 1B, 1C). To exclude that remaining flox and frt sites affected the proper expression of the Gfi1:Gfi1P2A knock-in allele, we prepared thymocytes from these animals, compared the length of wild type and knock-in transcript by RT-PCR and sequenced it (data not shown). Both transcripts had the same length and the knock in transcript had no other mutations except the P2A exchange indicating that the transcription of the Gfi1:Gfi1P2A allele was not disturbed (data not shown). Although it cannot be entirely ruled out that the remnant loxp and frt sites interfere with gene regulation, it is unlikely that they play a major role in our Gfi1:Gfi1P2A mutants since we detect the proper mRNA length and sequence. Co-Immunoprecipitation For coimmunoprecipitation, MEL 75A (clone 19) cells were used. After confluent growing, the cells were lysed and cell lysates (300µg) were incubated with anti-Gfi1b (Santa Cruz rabbbit polyclonal sc-22795) antibody overnight at 4 °C, followed by incubation with protein A/G Agarose (Roche) for an additional 2 h. The pellet was washed three times with TBSTween buffer and once with PBS. Then the pellet was resuspended in SDS-containing buffer, 1 Supplementary material, MS EMBO/2005/82698 third revision heated at 95 °C, and centrifuged at 12,000 g. The supernatant was subjected to SDS-PAGE. After transfer onto a nylon/nitrocellulose membrane, the samples were detected with either an anti-Gfi1b (Santa Cruz goat polyclonal antibody sc-8559) or an anti-PIAS3 antibody (Santa Cruz goat polyclonal sc-8154). For the input control, 15g protein was loaded on the gel. RT-PCR and Quantitative Real-Time PCR RNA from newborn inner ears was isolated using peqGOLD RNA pureTM (Peqlab). RNA was treated with DNaseI (Promega) for 30 min. Reverse Transcription was performed using Superscript II (Invitrogen) as described in the manufacturer`s protocol. For semiquantitative RT-PCR , the following primers were used 5’- CGCGGATCCATGCCACGGTCCTTTCTA-3’ and 5’-AGGTGTGTTTCTTCATGTCC-3’ for Gfi1b and 5’- GCGCTCATTCCTGGTCAAGA-3’ and 5’- TGTGTGGATGAAGGTGT GTT-3’ for amplification of Gfi1. Quantitative RT-PCR was performed in a 20 l reaction volume containing 900 nM of each primer, 250 nM TaqMan probe, and TaqMan Universal PCR Master Mix (Applied Biosystems). Reactions were monitored in an ABI PRISM 7700 Sequence Detection System (Applied Biosystems). For detection of Gfi1 (Mm 00515853_m1) and Gfi1b (Mm 00492319_m1) Assays on Demand from Applied Biosystems were used. Every PCR was performed in triplicates. 2 Supplementary material, MS EMBO/2005/82698 third revision Figure Legends Supp. Figure 1 Genotyping and expression analysis of Gfi1:Gfi1b knock-in mice. Heterozygous Gfi1:Gfi1b chimeras were born after injection of targeted ES clones in mouse blastocysts and were mated with CMV-Cre transgenic mice to delete the floxed neomycin selection cassette which was monitored by Southern blotting. A Southern blot analysis shows after digestion with BamHI and use of the 5' external probe an 8 kb fragment from the wild type allele and a 14 kb fragment from the targeted allele (left). Loss of the neomycin resistance gene can be detected using a SalI/SpeI double digestion and the 3' probe. This leads to an 11.9 kb fragment from the wild type allele and a 5.6 kb fragment from the targeted allele unless the neomycin resistance gene was deleted. If it is deleted, a 4.2 kb fragment is visible (middle and right). B Upper row: Gfi1 protein expression is detected in thymocytes from Gfi1+/+ and Gfi1+/1b mice. Gfi11b/1b thymocytes have lost Gfi1 expression. Detection of -actin was used as loading control. No expression of Gfi1 was detectable in thymocytes of homozygous Gfi1:Gfi1b (Gfi11b/1b) knock-in mouse mutants, which was expected since homozygosity for this allele disrupts the Gfi1 coding region. In contrast, Gfi1 expression is readily detected in heterozygous Gfi1:Gfi1b knock-ins (Gfi1+/1b) C Detection of Gfi1b expression in thymocytes from Gfi1+/1b and Gfi11b/1b mice. Expression of Gfi1b was undetectable in thymocytes of wild type animals (Gfi1+/+), as expected, but was clearly present in Gfi1+/1b and at a significantly higher level also in Gfi11b/1b animals. While these results confirm that functional Gfi1b alleles have been generated at the Gfi1 locus, the relatively increased Gfi1b protein level in Gfi11b/1b thymocytes indicates a regulatory phenomenon. It has been demonstrated that Gfi1 and Gfi1b are able to repress their own promoters and in some cells can also mutually down modulate their expression by using the same DNA binding sites in the Gfi1 and Gfi1b promoters, respectively (Doan et al., 2004, Yücel et al., 2004, Vassen et al., 2005). It is therefore likely that the presence of one Gfi1 and one Gfi1b allele in heterozygous animals results in different levels of expression than in homozygous or wild type mice. This seemed to be the case at least for Gfi1b expression in Gfi11b/1b cells. 3 Supplementary material, MS EMBO/2005/82698 third revision Supp. Figure 2 A Genotyping of Gfi1:Gfi1P2A knock-in mice. Southern blot of genomic DNA from ES cells (left) and mice (right) after HindIII digestion. DNA fragments with a size of 15 kb originate from the wild type allele while the targeted allele produces an 8 kb fragment. B Upper panel: Expression analysis by Western blot of whole cell extracts of thymocytes from Gfi1+/+, Gfi1+/P2A, Gfi1P2A/P2A and Gfi1-/- mice. Lower panel: Uniform loading is demonstrated by -actin expression level. 4 Supplementary material, MS EMBO/2005/82698 third revision Supp. Figure 3 A Expression analysis of Gfi1:Gfi1b knock-in mice. Semi-quantitative PCR from Gfi1+/+ and Gfi11b/1b inner ears of newborn mice. Upper row: Gfi1 mRNA is not detectable in Gfi11b/1b inner ears. PCR with Gfi1b specific primers shows a high mRNA expression in Gfi11b/1b inner ears compared to wild type (Gfi1+/+) mice. Lower panel: Uniform loading of RNA is demonstrated by GAPDH- expression level. B Expression analysis by quantitative Real Time PCR from wild type (left) and Gfi11b/1b (right) inner ears. PCR was performed with cDNA from newborn mice. The pictures show a weak expression of Gfi1 and Gfi1b in Gfi1+/+ inner ears. In Gfi11b/1b inner ears no Gfi1 cDNA can be detected, whereas the amount of Gfi1b cDNA is clearly increased in Gfi11b/1b inner ears compared to the wild type inner ears. 5 Supplementary material, MS EMBO/2005/82698 third revision Supp. Figure 4 Lack of interaction between Gfi1b and PIAS3 Upper panel: Shown is the detection of PIAS3 in a Western Blot with MEL75A cell extracts (input lane). MEL cell lysates precipitated with an anti Gfi1b antibody or an irrelevant anti GST antibody were probed with an anti PIAS3 antibody but no Gfi1b/PIAS3 complexes could be collected. As a control, the precipitates contain substantial amounts of Gfi1b when probed with an anti Gfi1b antibody (lower panel) indicating a properly executed immunoprecipitation reaction. Leftmost lane (lower panel) endogenous expression level of Gfi1b in MEL75A cells. 6 Supplementary material, MS EMBO/2005/82698 third revision Table Legend Table 1 Differential cell counts of wild type, Gfi1-/- , Gfi1P2A/P2A and Gfi11b/1b mice. Differential cell counts from May-Grünwald-Giemsa stained blood smears and bone marrow cytospins (after erythrocyte lysis). Shown are the mean values, their standard deviations and the results of a two tail students t-test. Each value obtained from mutant mice was compared to values obtained from wildtype animals. Significant results (p≤0.05) are marked with an asterisk. 7