* Your assessment is very important for improving the workof artificial intelligence, which forms the content of this project
Download Statistical Analyses of Microarray Data
Survey
Document related concepts
Endogenous retrovirus wikipedia , lookup
RNA silencing wikipedia , lookup
Secreted frizzled-related protein 1 wikipedia , lookup
Vectors in gene therapy wikipedia , lookup
Silencer (genetics) wikipedia , lookup
Gene therapy of the human retina wikipedia , lookup
Oligonucleotide synthesis wikipedia , lookup
Community fingerprinting wikipedia , lookup
Gene regulatory network wikipedia , lookup
Gene expression wikipedia , lookup
SNP genotyping wikipedia , lookup
Artificial gene synthesis wikipedia , lookup
Expression vector wikipedia , lookup
Virtual karyotype wikipedia , lookup
Transcript
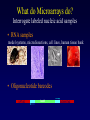
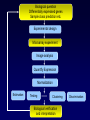
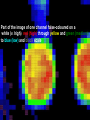
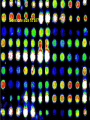
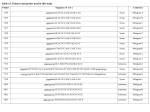
Statistical Analyses of Microarray Data Rafael A. Irizarry Department of Biostatistics [email protected] http://biosun01.biostat.jhsph.edu/~ririzarr Outline • • • • Scientific questions Review of technology Role of statistics Two case studies Scientific Questions • Expression “To understand gene function, it is helpful to know when and where it is expressed and…” • Differential expression “…under what circumstances the expression level is affected.” • Expression patterns “… questions concerning functional pathways and how cellular components work together to regulate and carry out cellular processes.” Lipshutz et al. (1999) Nature genetics, 21, pp. 20-21 What do Microarrays do? Interrogate labeled nucleic acid samples • RNA samples model systems, microdissections, cell lines, human tissue bank • Oligonucleotide barcodes UPTAG kanR DOWNTAG How do they do it? Labeled targets Probes cDNA Arrays cDNA clones (probes) excitation laser 2 scanning laser 1 emission PCR product amplification purification printing mRNA target overlay image and normalize 0.1nl/spot microarray Hybridize target to microarray analysis High Density Oligonucleotide Arrays GeneChip Probe Array Hybridized Probe Cell Single stranded, labeled RNA target * * * * * Oligonucleotide probe 24µm 1.28cm Millions of copies of a specific oligonucleotide probe >200,000 different complementary probes Image of Hybridized Probe Array Compliments of D. Gerhold Role of Statistics Biological question Differentially expressed genes Sample class prediction etc. Experimental design Microarray experiment Image analysis Quantify Expression Normalization Estimation Testing Clustering Biological verification and interpretation Discrimination Part of the image of one channel false-coloured on a white (v. high) red (high) through yellow and green (medium) to blue (low) and black scale Does one size fit all? Segmentation: limitation of the fixed circle method SRG Fixed Circle Inside the boundary is spot (fg), outside is not. Some local backgrounds Single channel grey scale We use something different again: a smaller, less variable value. Quantification of Expression For each spot on the slide we calculate Red intensity = Rfg – Rbg fg = foreground, bg = background, and Green intensity = Gfg – Gbg and combine them in the log (base 2) ratio Log2( Red intensity / Green intensity) we now have one differential expression for each gene for each array The red-green ratios can be spatially biased Top 2.5%of ratios red, bottom 2.5% of ratios green Another example Oligo Array Image Analysis • About 100 pixels per probe cell • These intensities are combined to form one number representing expression for the probe cell oligo Normalization at Probe Level Normalization at Probe Level Dilution Experiment Data Dilution Experiment Data PM MM Default until 2002 • GeneChip® software uses Avg.diff 1 Avg.diff ( PM j j MM j ) with A a set of “suitable” pairs chosen by software. • Log ratio version is also used. • For differential expression Avg.diffs are compared between chips. What is the evidence? Lockhart et. al. Nature Biotechnology 14 (1996) Two case studies Spike-In Experiments • Add concentrations (0.5pM – 100 pM) of 11 foreign species cRNAs to hybridization mixture • Set A: 11 control cRNAs were spiked in, all at the same concentration, which varied across chips. • Set B: 11 control cRNAs were spiked in, all at different concentrations, which varied across chips. The concentrations were arranged in 12x12 cyclic Latin square (with 3 replicates) Set A: Probe Level Data (12 chips) Spike-In B Probe Set Conc 1 Conc 2 Rank BioB-5 100 0.5 1 BioB-3 0.5 25.0 2 BioC-5 2.0 75.0 3 BioB-M 1.0 35.7 4 BioDn-3 1.5 50.0 5 DapX-3 35.7 3.0 6 CreX-3 50.0 5.0 7 CreX-5 12.5 2.0 8 BioC-3 25.0 100 9 DapX-5 5.0 1.5 10 DapX-M 3.0 1.0 11 Later we consider 23 different combinations of concentrations Observed Ranks Gene AvDiff MAS 5.0 Li&Wong AvLog(PM-BG) BioB-5 6 2 77 1 BioB-3 16 1 33 2 BioC-5 74 6 22 5 BioB-M 30 3 6 3 BioDn-3 44 5 27 4 DapX-3 239 24 796 7 CreX-3 333 73 386 11 CreX-5 3276 33 43 9 BioC-3 2709 8572 12 10300 DapX-5 2709 102 59 17 DapX-M 165 19 30 6 A B DOWNTAG kanR UPTAG CEN/ARS CEN/ARS Circular pRS416 EcoRI linearized PRS416 Transformation into deletion pool Select for Ura+ transformants Genomic DNA preparation Cy5 labeled PCR products PCR Cy3 labeled PCR products Oligonucleotide array hybridization NHEJ Defective YKU70 NEJ1 YKU80 YKU70 NEJ1 YKU80 • . Average Red and Green Scatter Plot Average Red and Green MVA plot Histograms QQ-Plot Z-Scores Average Red and Green MVA Plot Average Red and Green Scatter Plot Summary • Simple data exploration useful tool for quality assessment • Statistical thinking helpful for interpretation • Statistical models may help find signals in noise Acknowledgements Biostatistics Karl Broman Leslie Cope Carlo Coulantoni Giovanni Parmigiani Scott Zeger MBG (SOM) Jef Boeke Siew-Loon Ooi Marina Lee Forrest Spencer UC Berkeley Stat Ben Bolstad Sandrine Dudoit Terry Speed Jean Yang Gene Logic WEHI Francois Colin Bridget Hobbs Uwe Scherf’s Group Natalie Thorne PGA Tom Cappola Skip Garcia Joshua Hare